Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102751 |
| Name | oriT_pRphaR723d |
| Organism | Rhizobium phaseoli strain R723 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP013531 (955966..955994 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRphaR723d
AGGGCGCAATATACGTCGCTGCCGCGACG
AGGGCGCAATATACGTCGCTGCCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file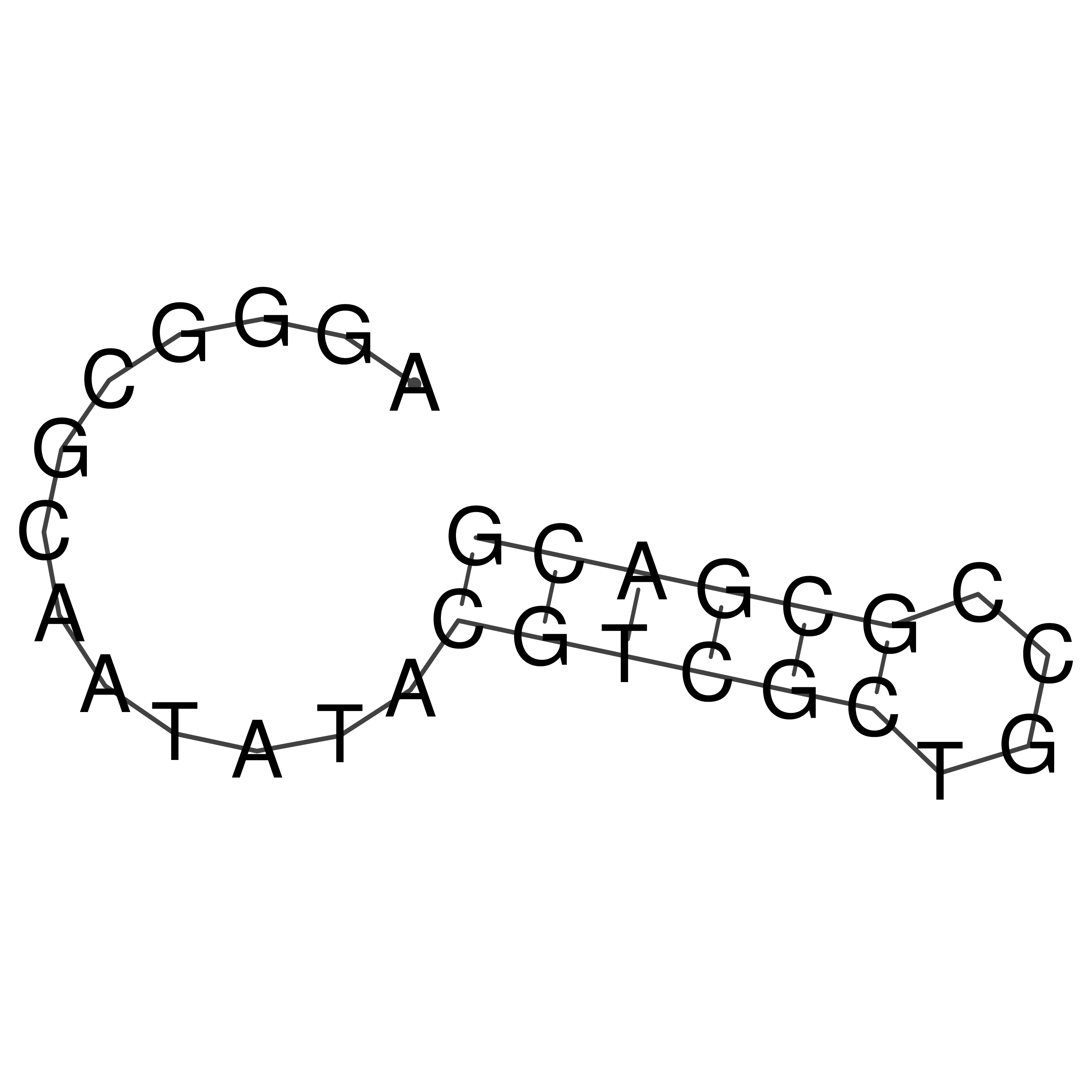
Relaxase
| ID | 2025 | GenBank | WP_064820496 |
| Name | traA_AMC89_RS31375_pRphaR723d |
UniProt ID | A0A7X6J0B9 |
| Length | 1543 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1543 a.a. Molecular weight: 171090.63 Da Isoelectric Point: 7.0921
>WP_064820496.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGISFSYRGGASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELFFAPADLARRQEMADRLLAEPELLLKQ
LGNESSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPALFTTRDMLRIE
YYMADSARRLSNRSGYAVSRKHVAAAIERIESDDPKNLFRLDAEQGDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VERGLDAYAQQGHLVEAGTREEAINRIVVDWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVKRLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLMSVLLDNGRNVVFSEDSYRNVDHGYASTIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAVKEDFQVRSEWGRRLRADHAAGVTGELVETGEAKFRPDDEDADDSHYADIRTGDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYDEREVDRNVWRAKQIETADARQERIERERHRP
DVFGRLVERLARSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLIPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASYTGTEPRGLADNLAVAPDRLGRLRGSELIVEGRAARDERDVAMA
ALEELLPLVRGHATEFRRNAERFELREQARRASMSLSVPALSEQAMARLMEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELIRLFEAVKRFAEEQH
LAERRDRSKIVAAASAVPGAAPGKQTVTVLPMLAAVTEFKTSVDDEARSRAIAAPLYRQQRAALAEAARM
IWRDPAATVDNIESLLEKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARL
RSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEVKNNGRTRNAAAGSLDPSIHREFD
AVSRALDERFGRSAIVRGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRNRTVNRGRG
IEL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGISFSYRGGASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEHGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGRELFFAPADLARRQEMADRLLAEPELLLKQ
LGNESSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDADTGKVSEPALFTTRDMLRIE
YYMADSARRLSNRSGYAVSRKHVAAAIERIESDDPKNLFRLDAEQGDAIRHVTGDSGIAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VERGLDAYAQQGHLVEAGTREEAINRIVVDWTEARKQAIDRSTSEGRDGHLRGDELLVLAHTNADVKRLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLMSVLLDNGRNVVFSEDSYRNVDHGYASTIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAVKEDFQVRSEWGRRLRADHAAGVTGELVETGEAKFRPDDEDADDSHYADIRTGDGTVHRLWGVSLPKA
LEEAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYDEREVDRNVWRAKQIETADARQERIERERHRP
DVFGRLVERLARSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGLAIERERRVAYNEERSETMIGAISSEERYLIPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASYTGTEPRGLADNLAVAPDRLGRLRGSELIVEGRAARDERDVAMA
ALEELLPLVRGHATEFRRNAERFELREQARRASMSLSVPALSEQAMARLMEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELIRLFEAVKRFAEEQH
LAERRDRSKIVAAASAVPGAAPGKQTVTVLPMLAAVTEFKTSVDDEARSRAIAAPLYRQQRAALAEAARM
IWRDPAATVDNIESLLEKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARL
RSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEVKNNGRTRNAAAGSLDPSIHREFD
AVSRALDERFGRSAIVRGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRNRTVNRGRG
IEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1674 | GenBank | WP_064820498 |
| Name | traG_AMC89_RS31390_pRphaR723d |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70447.36 Da Isoelectric Point: 9.3885
>WP_064820498.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSASFAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVALSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHTLGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GKAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQVIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSASFAMREFARLGPSVQTGVITWDRALSYLDMVAVLGAVAGFLAVALSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHTLGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GKAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQVIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 967873..977429
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AMC89_RS31435 (AMC89_PD00904) | 963448..963630 | + | 183 | WP_012490320 | hypothetical protein | - |
| AMC89_RS31440 (AMC89_PD00905) | 963763..964800 | - | 1038 | WP_064820506 | toprim domain-containing protein | - |
| AMC89_RS31445 (AMC89_PD00906) | 965163..966353 | + | 1191 | WP_064820507 | DUF1173 domain-containing protein | - |
| AMC89_RS31450 (AMC89_PD00907) | 966796..967389 | + | 594 | WP_037146723 | DUF433 domain-containing protein | - |
| AMC89_RS31455 (AMC89_PD00908) | 967404..967829 | + | 426 | WP_237357452 | PIN domain-containing protein | - |
| AMC89_RS31460 (AMC89_PD00909) | 967873..968868 | - | 996 | WP_064820508 | P-type DNA transfer ATPase VirB11 | virB11 |
| AMC89_RS31465 (AMC89_PD00910) | 968877..970052 | - | 1176 | WP_064820509 | type IV secretion system protein VirB10 | virB10 |
| AMC89_RS31470 (AMC89_PD00911) | 970061..970915 | - | 855 | WP_037146719 | P-type conjugative transfer protein VirB9 | virB9 |
| AMC89_RS31475 (AMC89_PD00912) | 970912..971583 | - | 672 | WP_037146717 | virB8 family protein | virB8 |
| AMC89_RS31480 (AMC89_PD00913) | 971577..971864 | - | 288 | WP_016737562 | hypothetical protein | - |
| AMC89_RS31485 (AMC89_PD00914) | 971905..972837 | - | 933 | WP_046977230 | type IV secretion system protein | virB6 |
| AMC89_RS31490 (AMC89_PD00915) | 972840..973073 | - | 234 | WP_016735278 | EexN family lipoprotein | - |
| AMC89_RS31495 (AMC89_PD00916) | 973070..973771 | - | 702 | WP_016735279 | P-type DNA transfer protein VirB5 | virB5 |
| AMC89_RS31500 (AMC89_PD00917) | 973768..976134 | - | 2367 | WP_046977231 | VirB4 family type IV secretion system protein | virb4 |
| AMC89_RS31505 (AMC89_PD00918) | 976127..976465 | - | 339 | WP_064820510 | type IV secretion system protein VirB3 | virB3 |
| AMC89_RS31510 (AMC89_PD00919) | 976471..976770 | - | 300 | WP_064820511 | TrbC/VirB2 family protein | virB2 |
| AMC89_RS31515 (AMC89_PD00920) | 976767..977429 | - | 663 | WP_064820512 | transglycosylase SLT domain-containing protein | virB1 |
| AMC89_RS31520 (AMC89_PD00921) | 977433..977969 | - | 537 | WP_064820513 | hypothetical protein | - |
| AMC89_RS31525 (AMC89_PD00922) | 978068..978439 | + | 372 | WP_041684118 | hypothetical protein | - |
| AMC89_RS31530 (AMC89_PD00923) | 978792..979982 | - | 1191 | WP_064820514 | hypothetical protein | - |
| AMC89_RS31535 (AMC89_PD00924) | 980057..980713 | - | 657 | WP_064820515 | hypothetical protein | - |
Host bacterium
| ID | 3194 | GenBank | NZ_CP013531 |
| Plasmid name | pRphaR723d | Incompatibility group | - |
| Plasmid size | 1074795 bp | Coordinate of oriT [Strand] | 955966..955994 [-] |
| Host baterium | Rhizobium phaseoli strain R723 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2 |
| Degradation gene | dxnH, LinG |
| Symbiosis gene | noeL, fixB, fixA |
| Anti-CRISPR | AcrIIA8 |