Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102724 |
| Name | oriT_pCFSAN004179G |
| Organism | Escherichia coli strain 08-00022 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP012501 (200541..200893 [+], 353 nt) |
| oriT length | 353 nt |
| IRs (inverted repeats) | 192..198, 206..212 (TATAAAA..TTTTATA) 197..202, 204..209 (AAAATA..TATTTT) 186..192, 196..202 (TATTTTT..AAAAATA) 104..110, 118..124 (TTTAAAT..ATTTAAA) 106..111, 113..118 (TAAATC..GATTTA) 93..98, 106..111 (GATTTA..TAAATC) 40..47, 50..57 (GCAAAAAC..GTTTTTGC) 4..11, 16..23 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 353 nt
>oriT_pCFSAN004179G
AGGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCGTTTAATATCATTGAGTTGTATTTGTGGATTTATATTGTTTAAATCTGATTTATTTAAAGCAGCGTCGTTAACGCAGCTACAGCAACGCGCCGACACCGCTTTGTAGGGGTGGTACTAACTATTTTTATAAAAAATATTATTTTATATTAGGGGGGGCTGCTAGCGGCGCGGTGTATTTTTTTATAGGATACCGCCAGGGGCGCTGCTAGCGGTGCGTCCCTGCTTGCATTATGAACTCTAGTGTTTCGAAATTAACTTTATTTTATGTTCAAAAAAGGTAATCTCTA
AGGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCGTTTAATATCATTGAGTTGTATTTGTGGATTTATATTGTTTAAATCTGATTTATTTAAAGCAGCGTCGTTAACGCAGCTACAGCAACGCGCCGACACCGCTTTGTAGGGGTGGTACTAACTATTTTTATAAAAAATATTATTTTATATTAGGGGGGGCTGCTAGCGGCGCGGTGTATTTTTTTATAGGATACCGCCAGGGGCGCTGCTAGCGGTGCGTCCCTGCTTGCATTATGAACTCTAGTGTTTCGAAATTAACTTTATTTTATGTTCAAAAAAGGTAATCTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file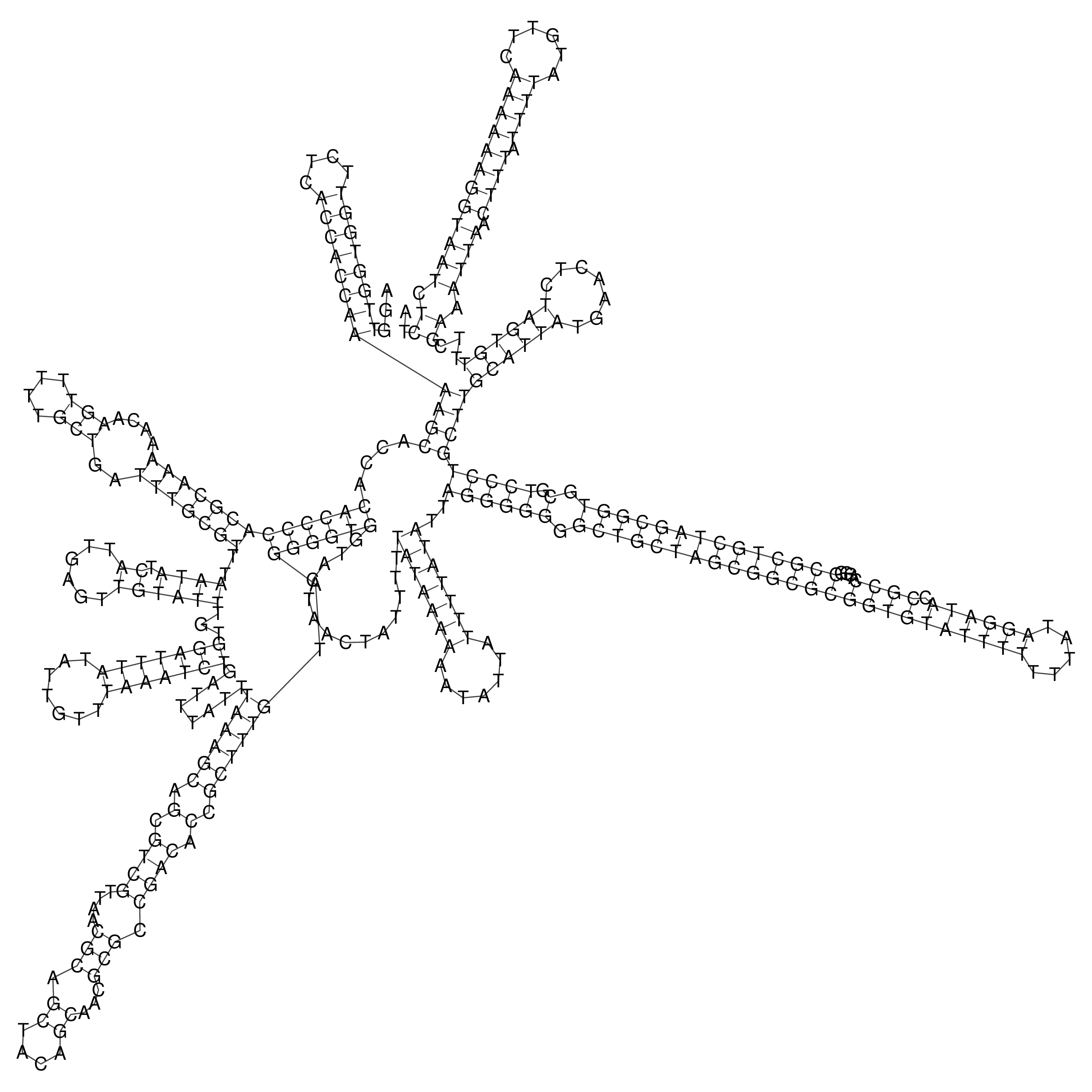
Auxiliary protein
| ID | 818 | GenBank | WP_001151539 |
| Name | WP_001151539_pCFSAN004179G |
UniProt ID | A0A0B1KNX7 |
| Length | 125 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 125 a.a. Molecular weight: 14339.42 Da Isoelectric Point: 5.3221
>WP_001151539.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Escherichia]
MAKVNLYISNDAYEKINVIIEKRRQEGAREKDVSFSATASMLLELGLRVYEAQMERKESAFNQIEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKND
MAKVNLYISNDAYEKINVIIEKRRQEGAREKDVSFSATASMLLELGLRVYEAQMERKESAFNQIEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKND
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0B1KNX7 |
T4CP
| ID | 1650 | GenBank | WP_047088906 |
| Name | traC_CP48_RS01260_pCFSAN004179G |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99295.98 Da Isoelectric Point: 5.9660
>WP_047088906.1 type IV secretion system protein TraC [Escherichia coli]
MNNPLEAVTQAVNSLLTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNFKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
MNNPLEAVTQAVNSLLTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNFKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1651 | GenBank | WP_047088736 |
| Name | traD_CP48_RS01390_pCFSAN004179G |
UniProt ID | _ |
| Length | 717 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 717 a.a. Molecular weight: 81410.98 Da Isoelectric Point: 5.0486
>WP_047088736.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Escherichia]
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKLSWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATIVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDLNPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGENVTQGEQPQQPASSVI
NDKKSDAGVSVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAVYEAWQREQNPDIQQQMQRCEEVN
INVHRERGEDVEPGDDF
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKLSWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATIVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDLNPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGENVTQGEQPQQPASSVI
NDKKSDAGVSVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAVYEAWQREQNPDIQQQMQRCEEVN
INVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 202872..231928
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CP48_RS01180 (CP48_p01325) | 198445..198732 | + | 288 | WP_000107542 | hypothetical protein | - |
| CP48_RS31165 (CP48_p01330) | 198851..199672 | + | 822 | Protein_240 | DUF932 domain-containing protein | - |
| CP48_RS30255 (CP48_p01340) | 199970..200479 | - | 510 | Protein_241 | transglycosylase SLT domain-containing protein | - |
| CP48_RS01205 (CP48_p01350) | 200894..201271 | + | 378 | WP_001151539 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| CP48_RS01210 (CP48_p01355) | 201471..202118 | + | 648 | WP_042966703 | transcriptional regulator TraJ family protein | - |
| CP48_RS01215 (CP48_p01360) | 202237..202464 | + | 228 | WP_000589558 | conjugal transfer relaxosome protein TraY | - |
| CP48_RS01220 (CP48_p01365) | 202498..202857 | + | 360 | WP_000340268 | type IV conjugative transfer system pilin TraA | - |
| CP48_RS01225 (CP48_p01370) | 202872..203183 | + | 312 | WP_000012108 | type IV conjugative transfer system protein TraL | traL |
| CP48_RS01230 (CP48_p01375) | 203205..203771 | + | 567 | WP_000399757 | type IV conjugative transfer system protein TraE | traE |
| CP48_RS01235 (CP48_p01380) | 203758..204486 | + | 729 | WP_001355329 | type-F conjugative transfer system secretin TraK | traK |
| CP48_RS01240 (CP48_p01385) | 204486..205904 | + | 1419 | WP_047088902 | F-type conjugal transfer pilus assembly protein TraB | traB |
| CP48_RS01245 (CP48_p01390) | 205894..206481 | + | 588 | WP_047088903 | conjugal transfer pilus-stabilizing protein TraP | - |
| CP48_RS01250 (CP48_p01395) | 206468..206788 | + | 321 | WP_047088904 | conjugal transfer protein TrbD | - |
| CP48_RS01255 (CP48_p01400) | 206785..207300 | + | 516 | WP_047088905 | type IV conjugative transfer system lipoprotein TraV | traV |
| CP48_RS28155 (CP48_p01405) | 207435..207656 | + | 222 | WP_042966711 | conjugal transfer protein TraR | - |
| CP48_RS01260 (CP48_p01415) | 207816..210443 | + | 2628 | WP_047088906 | type IV secretion system protein TraC | virb4 |
| CP48_RS01265 (CP48_p01420) | 210440..210826 | + | 387 | WP_047088907 | type-F conjugative transfer system protein TrbI | - |
| CP48_RS01270 (CP48_p01425) | 210823..211455 | + | 633 | WP_047088908 | type-F conjugative transfer system protein TraW | traW |
| CP48_RS01275 (CP48_p01430) | 211452..212444 | + | 993 | WP_047088909 | conjugal transfer pilus assembly protein TraU | traU |
| CP48_RS01280 (CP48_p01435) | 212760..214331 | - | 1572 | WP_000381395 | IS66-like element ISCro1 family transposase | - |
| CP48_RS01285 (CP48_p01440) | 214351..214698 | - | 348 | WP_000624622 | IS66 family insertion sequence element accessory protein TnpB | - |
| CP48_RS01290 (CP48_p01445) | 214698..215375 | - | 678 | WP_001339397 | IS66-like element accessory protein TnpA | - |
| CP48_RS01295 (CP48_p01450) | 215375..216091 | - | 717 | WP_065224995 | IS66-like element accessory protein TnpA | - |
| CP48_RS01300 (CP48_p01455) | 216170..216661 | + | 492 | WP_040091623 | hypothetical protein | - |
| CP48_RS01305 (CP48_p01460) | 216688..217326 | + | 639 | WP_040091622 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| CP48_RS28165 (CP48_p01465) | 217323..217703 | + | 381 | WP_024213661 | hypothetical protein | - |
| CP48_RS01310 (CP48_p01470) | 217938..219842 | + | 1905 | WP_236927347 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| CP48_RS01315 (CP48_p01475) | 219856..220113 | + | 258 | WP_040091333 | conjugal transfer protein TrbE | - |
| CP48_RS01320 (CP48_p01480) | 220106..220849 | + | 744 | WP_040091331 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| CP48_RS32860 | 220863..221226 | + | 364 | Protein_268 | hypothetical protein | - |
| CP48_RS01335 (CP48_p01500) | 221605..221886 | + | 282 | WP_000049684 | type-F conjugative transfer system pilin chaperone TraQ | - |
| CP48_RS01340 (CP48_p01505) | 221873..222409 | + | 537 | WP_040091329 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| CP48_RS01345 (CP48_p01510) | 222399..222713 | + | 315 | WP_040091328 | P-type conjugative transfer protein TrbJ | - |
| CP48_RS01350 (CP48_p01515) | 222667..223059 | + | 393 | WP_040091326 | F-type conjugal transfer protein TrbF | - |
| CP48_RS01355 (CP48_p01520) | 223046..224419 | + | 1374 | WP_024212142 | conjugal transfer pilus assembly protein TraH | traH |
| CP48_RS01360 (CP48_p01525) | 224416..227238 | + | 2823 | WP_047088737 | conjugal transfer mating-pair stabilization protein TraG | traG |
| CP48_RS01365 (CP48_p01530) | 227254..227751 | + | 498 | WP_040090738 | entry exclusion protein | - |
| CP48_RS01370 (CP48_p01535) | 227783..228514 | + | 732 | WP_040090739 | conjugal transfer complement resistance protein TraT | - |
| CP48_RS01375 (CP48_p01540) | 228574..228834 | + | 261 | WP_040090733 | hypothetical protein | - |
| CP48_RS30265 (CP48_p01545) | 229002..229719 | + | 718 | Protein_278 | hypothetical protein | - |
| CP48_RS01390 (CP48_p01550) | 229775..231928 | + | 2154 | WP_047088736 | type IV conjugative transfer system coupling protein TraD | virb4 |
| CP48_RS01395 (CP48_p01560) | 231937..232335 | - | 399 | WP_040090742 | type II toxin-antitoxin system VapC family toxin | - |
| CP48_RS01400 (CP48_p01565) | 232335..232562 | - | 228 | WP_000450526 | toxin-antitoxin system antitoxin VapB | - |
Host bacterium
| ID | 3167 | GenBank | NZ_CP012501 |
| Plasmid name | pCFSAN004179G | Incompatibility group | IncFIB |
| Plasmid size | 242187 bp | Coordinate of oriT [Strand] | 200541..200893 [+] |
| Host baterium | Escherichia coli strain 08-00022 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | estIa, faeC, faeD, faeE, faeF, faeH, faeI, faeJ, stcE, exeE, exeG, hlyD, hlyB, hlyA, hlyC |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7, AcrIF11 |