Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102666 |
| Name | oriT_GZWM139|p2 |
| Organism | Nitratireductor sp. GZWM139 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAMQML010000023 (32046..32145 [+], 100 nt) |
| oriT length | 100 nt |
| IRs (inverted repeats) | 24..31, 35..42 (TAATCGGC..GCCGATTA) |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 100 nt
GAATAAGGGAGAGTGAAGATAGTTAATCGGCTCCGCCGATTAGCTAACTTCACCTATCCTGCCCGCCTTACGGCGTTGTTTACACCAAGGAAAGTGTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file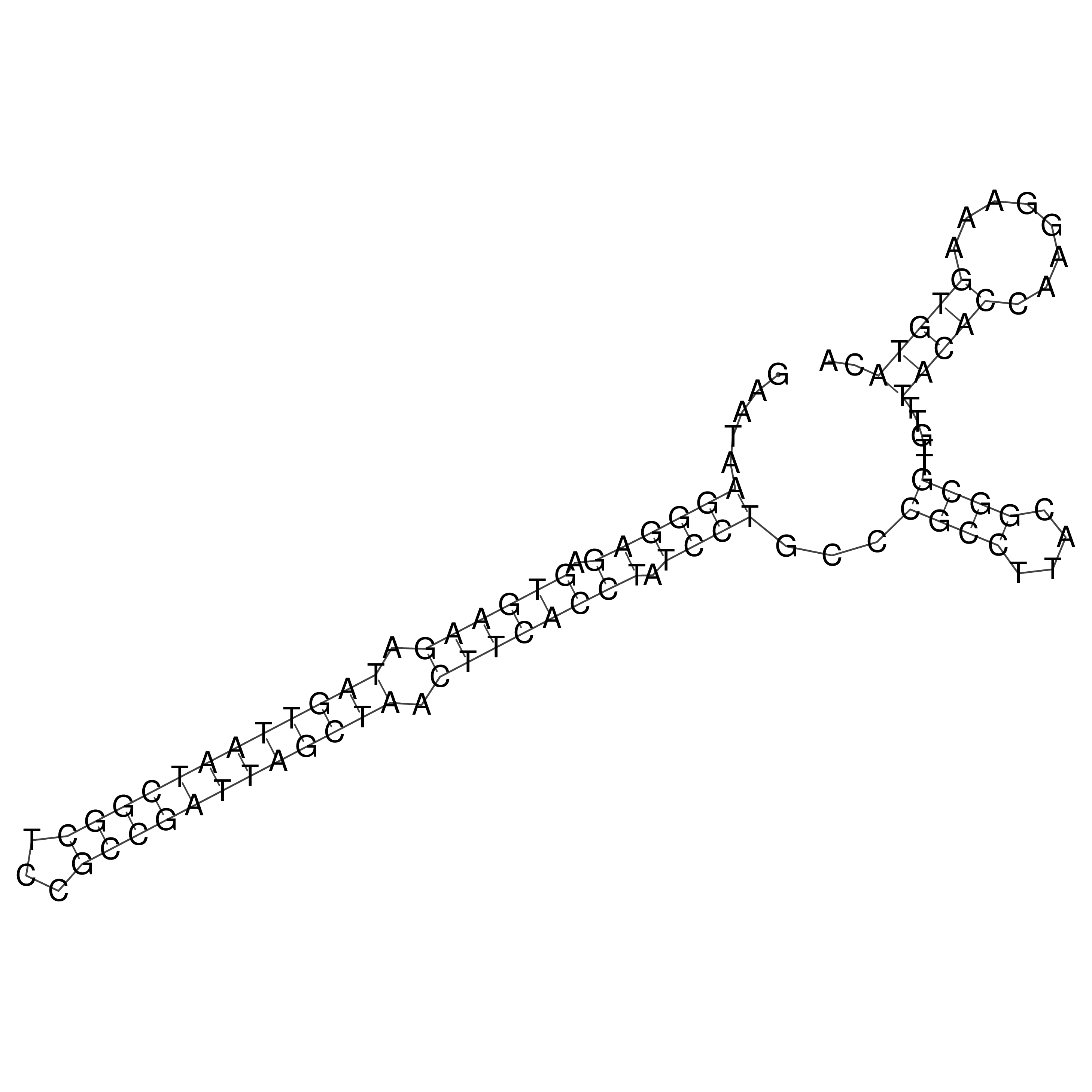
Relaxase
| ID | 1973 | GenBank | WP_283801641 |
| Name | Relaxase_NDI72_RS20575_GZWM139|p2 |
UniProt ID | _ |
| Length | 762 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 762 a.a. Molecular weight: 84222.45 Da Isoelectric Point: 10.5684
MIAKHVPMRSLKKSDFAGLVKYITDEQSKTERLGLVNATNCEADTMQAVIGEVLATQQGNTRAKGDKTYH
LIVSFPAGEKPEDDVLKAVEERICAGLGYADHQRVSAVHHDTDNVHIHIAINKIHPTRNTMHEPYQDYRT
LGGLCDALEDEYGLQKDNHQGRKSVSQGRASDMERHAGIESLVSWIKRECLEEIKTAKTWEELHQVMSDN
GLEIRQRANGFVIESEDGTQVKASTVARDLSKPKLEARLGAFEETAAQAERKAKRRYDKRPTHFKVNTTE
LYARYQEEQKNLTAARKVEWNNARGRKDRRIEAAKRSNRLRRSAIKLMGGSRISKKLLYAQAHKALKDEI
QSINKDYKREKQELYDQYKRRAWADWLKQQVLNGDKKALEALRAREQSKGLQGDTIKGEGGSKPGHAPVT
DNITKKGTIIFRTGKSAVRDDGDKLQVSKAANGDTVTEALRVAMERYGSRITVTGSPQFKAQVIVAAATS
NLPITFADAGLERRRQQLLQQENSNDRRNEQARRGNERGRADRRSNGRAERGPADDNGRGRGTADSRAAT
TGRAGDGQRGRGQSTSTGSTTAGGRNDAGNGGRPILNKPHVGRIGGEPPSFAKNRLRTLSSLGVVRFARG
GEVLLPGNAPGELEHQGAKPDNQLRWGVSGSGGITPDQTNAADKYIAEREAKRLKGFDIPKHSRYTNQDG
AVTYGGTRNVQGQALALLNRGDEVLVLPIDKATAQRMKRVRIGEAVTVTPQGSLKRSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 797 | GenBank | WP_269129272 |
| Name | WP_269129272_GZWM139|p2 |
UniProt ID | _ |
| Length | 123 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 123 a.a. Molecular weight: 13797.90 Da Isoelectric Point: 9.4637
MSEEKKPGKKRSHHLRVPVFDDEKETIEAQAARAGMSVARYLREVGQGYQIKGVVDYEQVRELARINGDL
GRLGGLLKLWLTDDVRTAQFGQSTILAVLSKIEATQEEMGKVMTKVVMPRSEP
Protein domains
No domain identified.
Protein structure
No available structure.
T4CP
| ID | 1609 | GenBank | WP_283801650 |
| Name | t4cp2_NDI72_RS20445_GZWM139|p2 |
UniProt ID | _ |
| Length | 845 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 845 a.a. Molecular weight: 94123.89 Da Isoelectric Point: 6.3396
MIQAISFAIAVCGAALLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAVVDDGVIVCKNGAFMASWL
YKGDDNASSTDEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERNTSAFPDPVSAAIDEERRQLF
EGLGAMYEGYFVLTLTWFPPLLAQRKFVELMFDDDAEVVGKKARTKGLINQFKREISSIENRLTTAVKLT
RLRGQKIVNEDGSKVTHDDFLSWLQFCVTGVSHPVQLPNNPMYMDAYIGGQEMWGGVVPKVGRKFIQVVS
IEGFPLESHPGILSALAELPVEYRWSSRFIFMDQHESVKHLDKFRKKWKQKVRGFFDQVFNTNSGNVDQD
ALTMVQDAESAIAEVNSGLVAVGYYTSVVVLMDESRDRLEQSARQVEKSINRLGFAARTETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGSADAPCPMYPPLSPALMHCVTSGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLALIAAQLRRYAGMSIFAFDKGMSMYPLTKAVGGLHFSVGADDDRLSFCPLQYLET
KSDRAWAMEWIDTILALNGVQSTPAQRNEIAHAIVSMHESGARTLSEFSLTIQDESIREAIRQYTVDGSM
GHLLDADEDGLSLTDFTTFEIEELMNLGEKFALPVLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRE
KIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMGLNARQIE
ILATAIPKRQYYYVSENGRRLYDLALGPLALSFVGASDKESVAAIKQLEAKYGEQWVHEWLAGRGLKLDN
YLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1610 | GenBank | WP_014579486 |
| Name | t4cp2_NDI72_RS20570_GZWM139|p2 |
UniProt ID | _ |
| Length | 635 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 635 a.a. Molecular weight: 69921.78 Da Isoelectric Point: 7.3667
MKPKINNAVGPQVRDRKPKKQRLLPALGAVSVLGGLQAATQFFAYTFNYQDNLGGHINHVYAPWSILSWA
SDWYSIYPDEFMRSGSIGMVVTTVGLLGVAIAKVVTSNSSKASEYLHGSARWAEKKDIQAAGLLPRERTA
LEVVTGKDAPTSTGVYVGGWEDKDGNFFYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWPQSSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSNGGVCWNPLDEIRIGTEYEVGDVQNLATLIVDPDGKGMDSHWQ
KTSYALLVGVILHALYKSKNEGTPATLPVVDAMLADPERDTGELWMEMATYGHVDGQNHPAVGSAARDMM
DRPEEEGGSVLSTAKSYLALYRDPVVARNVSKSEFRIKDLMNHDDPVSLYIVTQPNDKARLRPLVRVLTN
MIIRLLADEMSFEKGRPVAHYKHRLLAMLDEFPSLGKLEILQESLAFVAGYGIKCYLICQDLNQLKSRET
GYGPDETITSNCHVQNAYPPNRIETAEHLSRLTGQTTVVKEQITTSGRRTSAMLGQVSRTIQEVQRPLLT
PDECLRMPGPTKNAAGDIEKAGDMVIYVAGYPAVYGKQPLYFKDPVFQARASVDAPKISDKLRIVAEPEG
EGVTI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4148..14936
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NDI72_RS20415 (NDI72_20405) | 1782..2921 | - | 1140 | WP_283801649 | plasmid replication initiator TrfA | - |
| NDI72_RS20420 (NDI72_20410) | 3069..3413 | - | 345 | WP_283801628 | single-stranded DNA-binding protein | - |
| NDI72_RS20425 (NDI72_20415) | 3513..3896 | + | 384 | WP_041646689 | transcriptional regulator | - |
| NDI72_RS20430 (NDI72_20420) | 4148..5107 | + | 960 | WP_014579515 | P-type conjugative transfer ATPase TrbB | virB11 |
| NDI72_RS20435 (NDI72_20425) | 5133..5558 | + | 426 | WP_014579514 | conjugal transfer system pilin TrbC | virB2 |
| NDI72_RS20440 (NDI72_20430) | 5561..5872 | + | 312 | WP_008929954 | conjugal transfer protein TrbD | virB3 |
| NDI72_RS20445 (NDI72_20435) | 5872..8409 | + | 2538 | WP_283801650 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| NDI72_RS20450 (NDI72_20440) | 8409..9191 | + | 783 | WP_269129290 | conjugal transfer protein TrbF | virB8 |
| NDI72_RS20455 (NDI72_20445) | 9209..10114 | + | 906 | WP_283801629 | P-type conjugative transfer protein TrbG | virB9 |
| NDI72_RS20460 (NDI72_20450) | 10117..10626 | + | 510 | WP_283801630 | conjugal transfer protein TrbH | - |
| NDI72_RS20465 (NDI72_20455) | 10630..12057 | + | 1428 | WP_269129287 | TrbI/VirB10 family protein | virB10 |
| NDI72_RS20470 (NDI72_20460) | 12076..12837 | + | 762 | WP_269129286 | P-type conjugative transfer protein TrbJ | virB5 |
| NDI72_RS20475 (NDI72_20465) | 12847..13074 | + | 228 | WP_269129285 | entry exclusion lipoprotein TrbK | - |
| NDI72_RS20480 (NDI72_20470) | 13086..14936 | + | 1851 | WP_283801631 | P-type conjugative transfer protein TrbL | virB6 |
| NDI72_RS20485 (NDI72_20475) | 14956..15438 | + | 483 | WP_283801632 | hypothetical protein | - |
| NDI72_RS20490 (NDI72_20480) | 15450..16085 | + | 636 | WP_283801633 | muramidase | - |
| NDI72_RS20495 (NDI72_20485) | 16082..16810 | + | 729 | WP_283801634 | TraX family protein | - |
| NDI72_RS20500 (NDI72_20490) | 16824..17249 | + | 426 | WP_014579501 | OmpA family protein | - |
| NDI72_RS20505 (NDI72_20495) | 17268..17537 | + | 270 | WP_283801635 | hypothetical protein | - |
| NDI72_RS20510 (NDI72_20500) | 17562..17816 | + | 255 | WP_283801636 | hypothetical protein | - |
| NDI72_RS20515 (NDI72_20505) | 17821..18327 | + | 507 | WP_283801637 | hypothetical protein | - |
| NDI72_RS20520 (NDI72_20510) | 18412..18717 | + | 306 | WP_283801638 | hypothetical protein | - |
| NDI72_RS20525 (NDI72_20515) | 18738..19361 | - | 624 | WP_283801639 | recombinase family protein | - |
| NDI72_RS20530 | 19370..19501 | - | 132 | WP_269129278 | hypothetical protein | - |
Host bacterium
| ID | 3110 | GenBank | NZ_JAMQML010000023 |
| Plasmid name | GZWM139|p2 | Incompatibility group | - |
| Plasmid size | 42560 bp | Coordinate of oriT [Strand] | 32046..32145 [+] |
| Host baterium | Nitratireductor sp. GZWM139 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |