Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102586 |
| Name | oriT_pA12b |
| Organism | Rhizobium tropici strain A12 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAADZA010000042 (7581..7616 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 21..26, 31..36 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pA12b
ATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
ATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file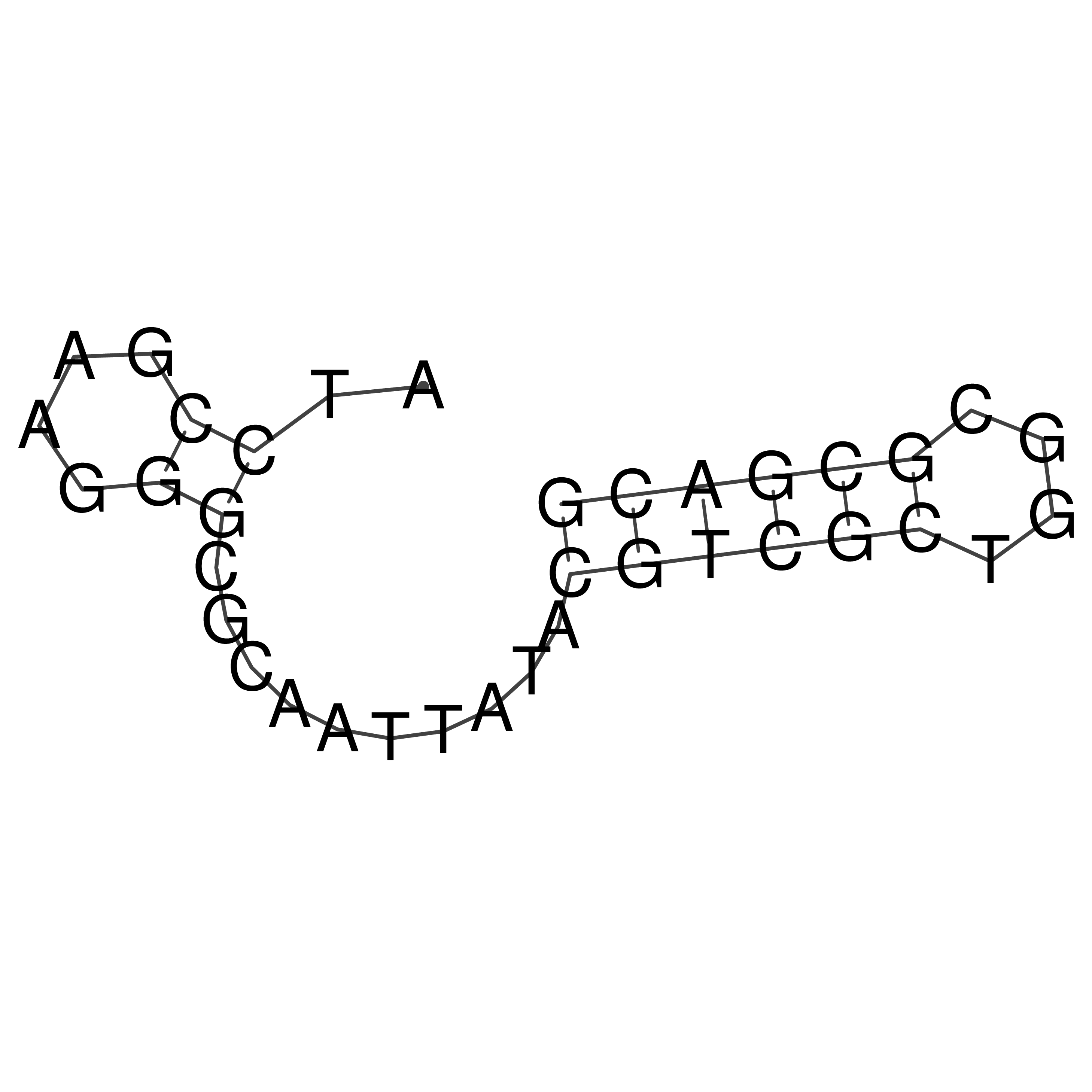
Relaxase
| ID | 1934 | GenBank | WP_004119830 |
| Name | traA_GXW80_RS25635_pA12b |
UniProt ID | A0A6P1CE10 |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122039.84 Da Isoelectric Point: 10.0272
>WP_004119830.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADAPSWLRTMIADR
SVSGASEAFWNSVEIFEKRVDAQLAKDVTVALPMELSSEQNIELVRDFIERHITSKGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPGGKPVRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLARAG
LDLRVDGHSFEKQGINLTPTIHLGVGAKAIERKSAEAERAPSLERLELQEERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQSLMARILQSPETLRLERERIEFSGGLRVPTKYTTRDLVRLEA
EMANRAIWLAQRSSHGVADAVREATFARHLRLADEQKTAIAHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYGELENIYRQREQWMREASLDLARGNIGRAVEAYR
AQGRVSGADLKADAVQALIADWNRDYDPQRSSLILAHLRRDVRMLNGMARAKLVERGLVADGFAFRTEDG
HRKFAAGDQIVFLKNEGSLGVKNGMLAKVVDATPGRIVAEVGDGEHRRQVTVEQRFYANLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDFGLYHGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
AQALRFADARGLHLVNVARTVVRDRIEWTVRQKSKLAALGARLAAIARRLGLSAGAAKTSQSHSMQEAKP
MVSGITTFPKSLEQVVEDLLAADAGLKKQWEEVSARFHLVYAQPEDAFKAVNVDAMLRNEAAAKSTITKI
GSEPETFGALKGKTGILAGRADKQAHERAVTNAPALARNLERYLRQRTEAEHKYDAEERAVRMKISLDIP
ALSPAAKQTLERVRDAIDRNDLPAGLENALADKMVKAELEGFARAVSERFGERTFLPLAAKDMTGQTFQS
VTSSMSAGQKAEVQSAWSTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADAPSWLRTMIADR
SVSGASEAFWNSVEIFEKRVDAQLAKDVTVALPMELSSEQNIELVRDFIERHITSKGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPGGKPVRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLARAG
LDLRVDGHSFEKQGINLTPTIHLGVGAKAIERKSAEAERAPSLERLELQEERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQSLMARILQSPETLRLERERIEFSGGLRVPTKYTTRDLVRLEA
EMANRAIWLAQRSSHGVADAVREATFARHLRLADEQKTAIAHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYGELENIYRQREQWMREASLDLARGNIGRAVEAYR
AQGRVSGADLKADAVQALIADWNRDYDPQRSSLILAHLRRDVRMLNGMARAKLVERGLVADGFAFRTEDG
HRKFAAGDQIVFLKNEGSLGVKNGMLAKVVDATPGRIVAEVGDGEHRRQVTVEQRFYANLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDFGLYHGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
AQALRFADARGLHLVNVARTVVRDRIEWTVRQKSKLAALGARLAAIARRLGLSAGAAKTSQSHSMQEAKP
MVSGITTFPKSLEQVVEDLLAADAGLKKQWEEVSARFHLVYAQPEDAFKAVNVDAMLRNEAAAKSTITKI
GSEPETFGALKGKTGILAGRADKQAHERAVTNAPALARNLERYLRQRTEAEHKYDAEERAVRMKISLDIP
ALSPAAKQTLERVRDAIDRNDLPAGLENALADKMVKAELEGFARAVSERFGERTFLPLAAKDMTGQTFQS
VTSSMSAGQKAEVQSAWSTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A6P1CE10 |
T4CP
| ID | 1564 | GenBank | WP_004119814 |
| Name | traG_GXW80_RS25650_pA12b |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 71115.08 Da Isoelectric Point: 9.0145
>WP_004119814.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTRILLFVAPCALMMLMAIGMTGTELWLSHFGKSDAARQMLGRAGIALPYVAASVVGIIFLFASAGSARI
RTAGWGVVTGATATLVLAVLREASRLSAYLEQVPAGKTVFNYLDPATAIGAAAVLMSALFGMRVAIAGNA
TFARAEPKRIVGKRALHGEADWMKLSQAEKLFAESGGIVIGERYRVDRDSIAAPSFRADSAASWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVLKHRGDANRDVFVLDPK
HSEIGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTTEND
QTLRQVRKNLSEPEPQLRQRLHEIYDDSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAAL
VSGTSFSTSDLADGKTDVFINIDLKTLETHAGLARVIIGAFLNAIYNRNGEMQGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKGCVGAR
AFSKAANPAENSPDRPARSAASKADREQ
MTRILLFVAPCALMMLMAIGMTGTELWLSHFGKSDAARQMLGRAGIALPYVAASVVGIIFLFASAGSARI
RTAGWGVVTGATATLVLAVLREASRLSAYLEQVPAGKTVFNYLDPATAIGAAAVLMSALFGMRVAIAGNA
TFARAEPKRIVGKRALHGEADWMKLSQAEKLFAESGGIVIGERYRVDRDSIAAPSFRADSAASWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVLKHRGDANRDVFVLDPK
HSEIGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTTEND
QTLRQVRKNLSEPEPQLRQRLHEIYDDSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAAL
VSGTSFSTSDLADGKTDVFINIDLKTLETHAGLARVIIGAFLNAIYNRNGEMQGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKGCVGAR
AFSKAANPAENSPDRPARSAASKADREQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 3030 | GenBank | NZ_JAADZA010000042 |
| Plasmid name | pA12b | Incompatibility group | - |
| Plasmid size | 45675 bp | Coordinate of oriT [Strand] | 7581..7616 [-] |
| Host baterium | Rhizobium tropici strain A12 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodM |
| Anti-CRISPR | - |