Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102584 |
| Name | oriT_pSM135A_Rh02 |
| Organism | Rhizobium leguminosarum strain SM135A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIOO01000003 (81373..81401 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSM135A_Rh02
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file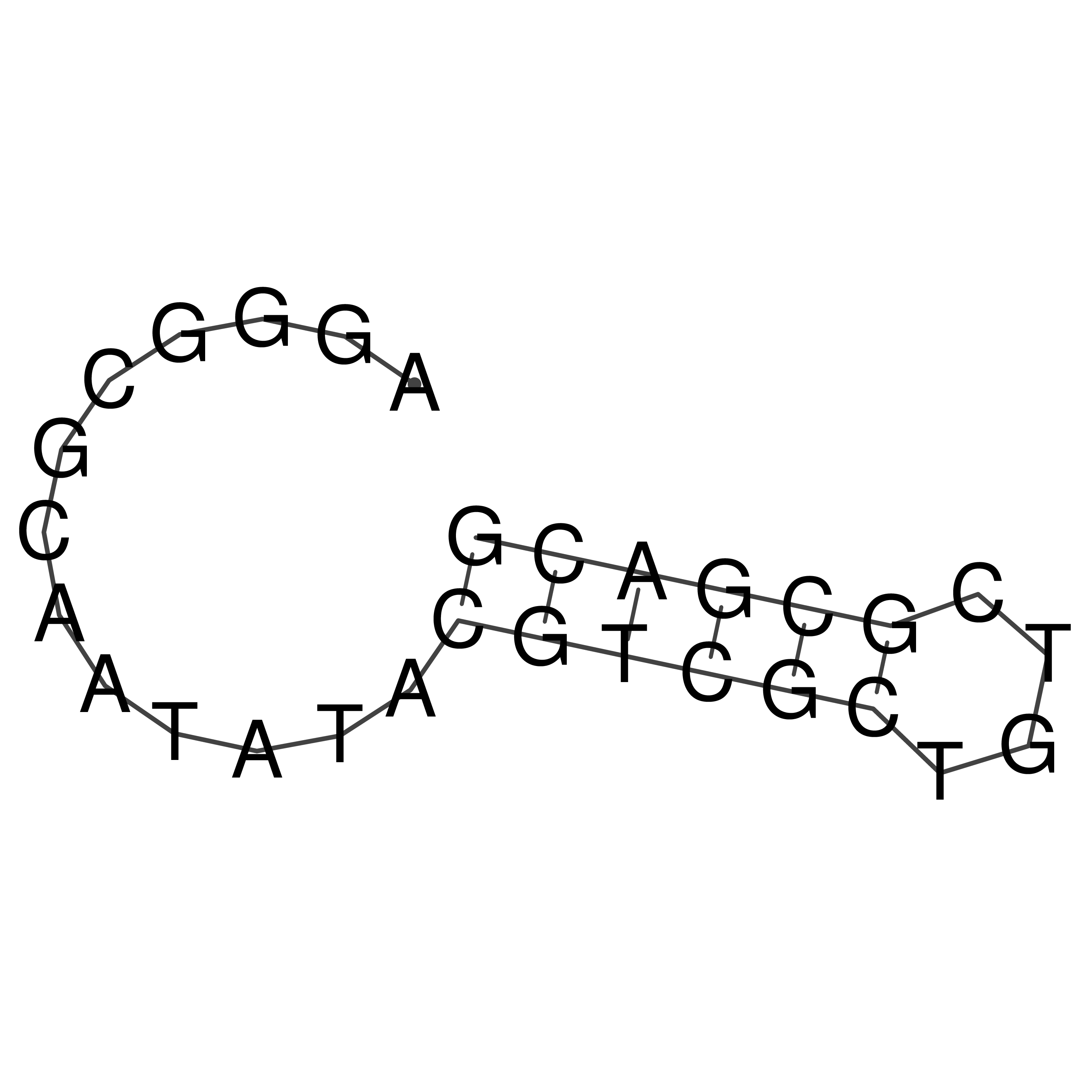
Relaxase
| ID | 1932 | GenBank | WP_130751505 |
| Name | traA_ELH89_RS30065_pSM135A_Rh02 |
UniProt ID | _ |
| Length | 1536 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1536 a.a. Molecular weight: 169711.00 Da Isoelectric Point: 8.6768
>WP_130751505.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAIMFVRAQVIGRGAGRSVVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQIPAWLRSAISG
QSVAKASETLWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVLDNLTAKGMVADWVYHDKDG
NPHIHLMTTLRPLAEEGFGPKKVPVRGEDGKPLRVVTPDRPNGKVVYKLWAGDKETLQGWKIAWAETASR
HLALAGHEIRLDGRSYAEQGLDGIAQRHLGPEKAALSRRDAEMYFAPADLARRQKMADRLLADPALLLKQ
LGNERSTFDERDIAKALHRTVDDPADFANIRARLMASDDLIMLKPQQIDAETGKAREPAVFTTREMLRIE
YDMGRSARVLAERRGFAVSDRVVAAAIRQVETQDPENSFRLDPEQVDAIHHITRDNAIAAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELAWASGRELLERDNVLVIDEAGMVS
SQQMARMLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQPWAREASRLFARGE
TGKGLDAYAQQGHLIEAGTRVETIDRIVADWSQARKQAIQNSTTAGNDGRLRGDELLVLAHTNEDVKYLN
EALRSVMVGEGALGEDRAFRTERGARAFAAGDRIIFLENARFVEPRATRLGPQHVKNGMLGTVVSTGDKR
GDAVLSVRLDSGRDVVISESSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKDDFEAKPEWGRKPHVDHAAGVTGALVETGQAKFRPNDEDADDSPYADVRADDGTLHRLWGVSLPKA
LEEGGVSEGDSVTLRKDGVERVTIKFAIVDEQTGQKRFEEREVDRNVWTARRIETAEARQQRIERESHRP
DVFKQLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAGVEESLSRRWAGIAEKREQLA
KLWERASVALGFTIEREPRIAYNEERAEPQTVLEADVARGRYLIPPTTGFARSAEEDARLAQLSSPAWKE
REAILRPLLETIYRDPDAALVALNALSSDTSVDPRRLADELAKAPERLGRLRGSDLIVDGRAARDERNAA
ITALTDLLPLVRAHATQFRRNAERFESHEQQRRAHMALPIPALSEKVMARLTEIEAVRRQGGDDAYKTAF
ALAAKDRTIVQEVKAVSEALTARFGWSAFTAKADAVGERNMVERMPEDLASIRGEKLTRLFKAVKRFADE
QHLAERRDRSKIVAGASADQGMENVPMLAAVTEFRTAIDEEARSRVLSDPLYRQQRAALAAVASAIWRDP
VGAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKEAVQAVPEAAARLRSLGP
AYVNVLDAERQAIAEERRRMAVAIPALSTAAEDVLMRLTAEAKNNDRKLTASAAPLDPSIRREFAAVSRA
LDERFGRNAIIRGEKDLINRVPPAQRNAFEAMQDRLKVLQRTVRMESSEQIISERRQRAVSRGISL
MAIMFVRAQVIGRGAGRSVVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQIPAWLRSAISG
QSVAKASETLWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVLDNLTAKGMVADWVYHDKDG
NPHIHLMTTLRPLAEEGFGPKKVPVRGEDGKPLRVVTPDRPNGKVVYKLWAGDKETLQGWKIAWAETASR
HLALAGHEIRLDGRSYAEQGLDGIAQRHLGPEKAALSRRDAEMYFAPADLARRQKMADRLLADPALLLKQ
LGNERSTFDERDIAKALHRTVDDPADFANIRARLMASDDLIMLKPQQIDAETGKAREPAVFTTREMLRIE
YDMGRSARVLAERRGFAVSDRVVAAAIRQVETQDPENSFRLDPEQVDAIHHITRDNAIAAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELAWASGRELLERDNVLVIDEAGMVS
SQQMARMLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQPWAREASRLFARGE
TGKGLDAYAQQGHLIEAGTRVETIDRIVADWSQARKQAIQNSTTAGNDGRLRGDELLVLAHTNEDVKYLN
EALRSVMVGEGALGEDRAFRTERGARAFAAGDRIIFLENARFVEPRATRLGPQHVKNGMLGTVVSTGDKR
GDAVLSVRLDSGRDVVISESSYRNVDHGYAATIHKSQGATVERTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKDDFEAKPEWGRKPHVDHAAGVTGALVETGQAKFRPNDEDADDSPYADVRADDGTLHRLWGVSLPKA
LEEGGVSEGDSVTLRKDGVERVTIKFAIVDEQTGQKRFEEREVDRNVWTARRIETAEARQQRIERESHRP
DVFKQLVERLSRSGAKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAGVEESLSRRWAGIAEKREQLA
KLWERASVALGFTIEREPRIAYNEERAEPQTVLEADVARGRYLIPPTTGFARSAEEDARLAQLSSPAWKE
REAILRPLLETIYRDPDAALVALNALSSDTSVDPRRLADELAKAPERLGRLRGSDLIVDGRAARDERNAA
ITALTDLLPLVRAHATQFRRNAERFESHEQQRRAHMALPIPALSEKVMARLTEIEAVRRQGGDDAYKTAF
ALAAKDRTIVQEVKAVSEALTARFGWSAFTAKADAVGERNMVERMPEDLASIRGEKLTRLFKAVKRFADE
QHLAERRDRSKIVAGASADQGMENVPMLAAVTEFRTAIDEEARSRVLSDPLYRQQRAALAAVASAIWRDP
VGAVGKIEELLQKGFAGERIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKEAVQAVPEAAARLRSLGP
AYVNVLDAERQAIAEERRRMAVAIPALSTAAEDVLMRLTAEAKNNDRKLTASAAPLDPSIRREFAAVSRA
LDERFGRNAIIRGEKDLINRVPPAQRNAFEAMQDRLKVLQRTVRMESSEQIISERRQRAVSRGISL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 3028 | GenBank | NZ_SIOO01000003 |
| Plasmid name | pSM135A_Rh02 | Incompatibility group | - |
| Plasmid size | 618182 bp | Coordinate of oriT [Strand] | 81373..81401 [+] |
| Host baterium | Rhizobium leguminosarum strain SM135A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |