Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102582 |
| Name | oriT_pSM135B_Rh04 |
| Organism | Rhizobium leguminosarum strain SM135B |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIOP01000004 (349132..349167 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 15..21, 26..32 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pSM135B_Rh04
AAGGGCGCAATTATACGTCGCTGACGCGACGTCTTG
AAGGGCGCAATTATACGTCGCTGACGCGACGTCTTG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file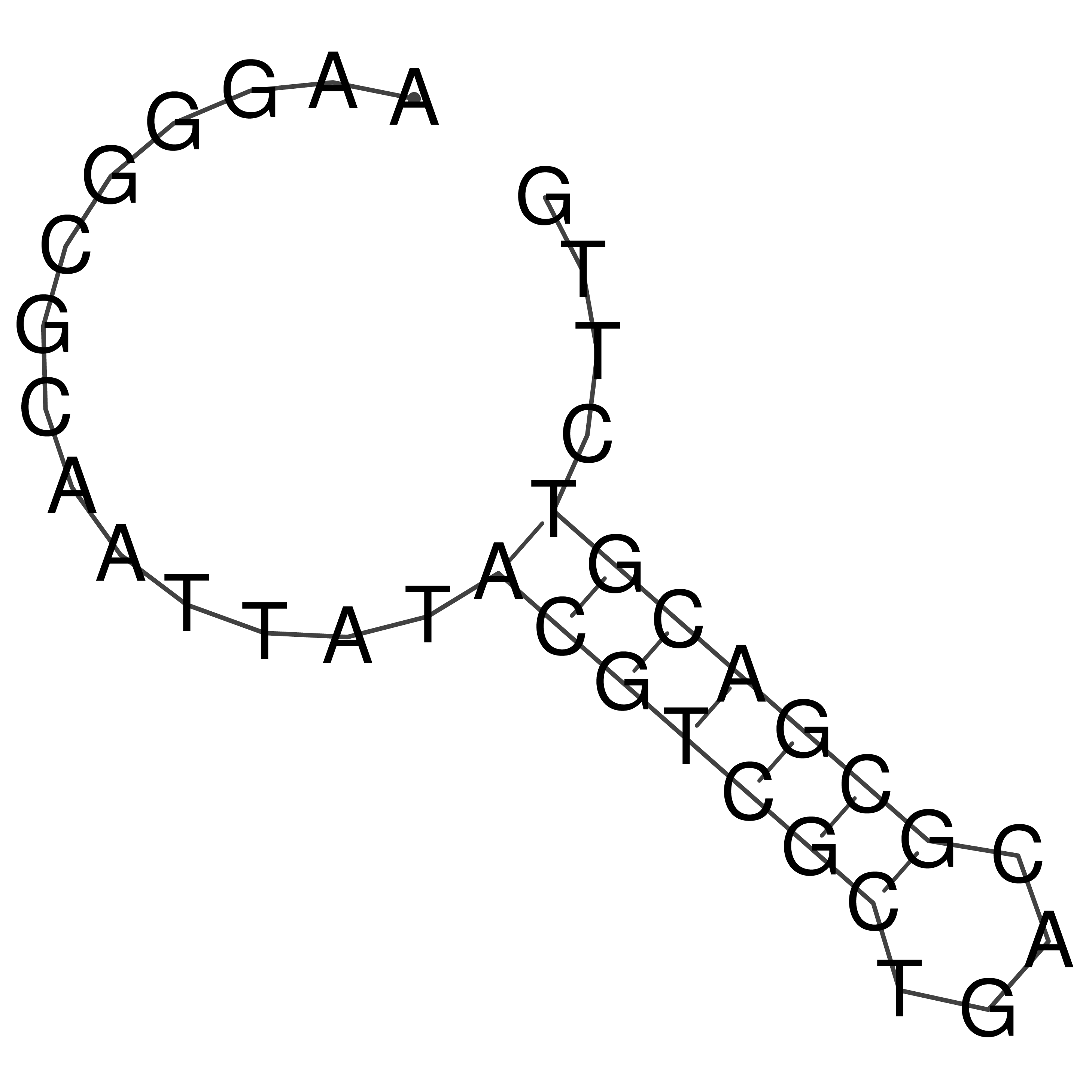
Relaxase
| ID | 1930 | GenBank | WP_130719407 |
| Name | traA_ELH90_RS35020_pSM135B_Rh04 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123062.99 Da Isoelectric Point: 9.7536
>WP_130719407.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARKIDYTRKRGLLHEEFVIPVDAPQWLRTMIADR
SVSGASEAFWNRVEAFEKRSDAQLAKDVTIALPIELSAGQNIALVRDFVGRHITANGMVADWVYHDAAGN
PHVHLMTTLRPLTEDGFGAKKVAVLGPDGNLLRNDSGKILYEAWAGSVDDFHVFRDGWFACQNEHLALAG
LDIRIDGRSFEAQGVELTPTIHLGVGTKAIERKAEQAKAKVSVWALKLERIELQEDRRVENARRIQRRPE
IVLDLITREKSVFDERDVAKILHRYIDDTGLFQSLMVRILQSPETLPLERERIEFGTGIRTPAKYTTRDL
IRLEADMTNRAIWLAARNSHGVKKSVLEATFAHHSRLSQEQRSAIAHLAATEQVAAVVGRAGAGKTTMMT
AAREAWESSGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELDWNEGRNQLDGRTVFVLDEAGMVSSRQM
AVFVEAASKAGAKLVLIGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREHWMRDASLDLARGNIATA
VDAYRANDRLLGSELKAHAVENLIADWNRDYDPTKTTLILAHLRRDVRMLNELARENLVERGIVATGFAF
RTEEGNRNFAAGDQIVFLRNEGSLGVKNGMIGKVVEAAPCRILVEVGQDEHRRQVTVDQRFYNNVDHGYA
ITIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHRDDLAVYYGSRSFAKAGGLIPILSRKNAKEMTLDYE
KNSLYRQALRFSEARGLHLMNVARTIARDRLDWTVRQKQKLADLGARLLAIGSMLGVADRIAKSSLPNAI
KESKPMVSGISTFPKSIEQVVEDKLAADAGLRTQWEDVSTRFHLVYAQPEAAFRAVDVDAMLKDQSTAKA
TLAKIAAEPASFGALRGKTGIFANRSDKHDRDRAIANGPALARNLERYLRQRTEAERKYDAEERGVRLKI
SIDIPALSPAARQTLERVRDAIDRNDLAAGFEYALADKLVNAELEGLAKAVSERFGERTFLGVASKDASG
GTYKSITSGMNPTQKAEMRIAWDWIRTIQQLSAHRNTTEALKQAQTVRQTKGPGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARKIDYTRKRGLLHEEFVIPVDAPQWLRTMIADR
SVSGASEAFWNRVEAFEKRSDAQLAKDVTIALPIELSAGQNIALVRDFVGRHITANGMVADWVYHDAAGN
PHVHLMTTLRPLTEDGFGAKKVAVLGPDGNLLRNDSGKILYEAWAGSVDDFHVFRDGWFACQNEHLALAG
LDIRIDGRSFEAQGVELTPTIHLGVGTKAIERKAEQAKAKVSVWALKLERIELQEDRRVENARRIQRRPE
IVLDLITREKSVFDERDVAKILHRYIDDTGLFQSLMVRILQSPETLPLERERIEFGTGIRTPAKYTTRDL
IRLEADMTNRAIWLAARNSHGVKKSVLEATFAHHSRLSQEQRSAIAHLAATEQVAAVVGRAGAGKTTMMT
AAREAWESSGYRVVGGALAGKAAEGLEKEAGIQSRTLSSWELDWNEGRNQLDGRTVFVLDEAGMVSSRQM
AVFVEAASKAGAKLVLIGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREHWMRDASLDLARGNIATA
VDAYRANDRLLGSELKAHAVENLIADWNRDYDPTKTTLILAHLRRDVRMLNELARENLVERGIVATGFAF
RTEEGNRNFAAGDQIVFLRNEGSLGVKNGMIGKVVEAAPCRILVEVGQDEHRRQVTVDQRFYNNVDHGYA
ITIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHRDDLAVYYGSRSFAKAGGLIPILSRKNAKEMTLDYE
KNSLYRQALRFSEARGLHLMNVARTIARDRLDWTVRQKQKLADLGARLLAIGSMLGVADRIAKSSLPNAI
KESKPMVSGISTFPKSIEQVVEDKLAADAGLRTQWEDVSTRFHLVYAQPEAAFRAVDVDAMLKDQSTAKA
TLAKIAAEPASFGALRGKTGIFANRSDKHDRDRAIANGPALARNLERYLRQRTEAERKYDAEERGVRLKI
SIDIPALSPAARQTLERVRDAIDRNDLAAGFEYALADKLVNAELEGLAKAVSERFGERTFLGVASKDASG
GTYKSITSGMNPTQKAEMRIAWDWIRTIQQLSAHRNTTEALKQAQTVRQTKGPGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1558 | GenBank | WP_130719405 |
| Name | traG_ELH90_RS35005_pSM135B_Rh04 |
UniProt ID | _ |
| Length | 682 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 682 a.a. Molecular weight: 74201.75 Da Isoelectric Point: 9.3084
>WP_130719405.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTANKIALVGLPITLMILVTIGMTGSEQWLSAFGKTEAGRRMLGRAGIALPYIASATVGMIVLFASAGST
NIKLAGWSVVGGSSVTILIAALRETVRLVALLDQLPTARSALFYADPATMIGAAAALISAAFALRVSLVG
NAAFATAEPKRILGRRALHGEADWMKMTQAGKLFPDAGGIVIGERYRVDKDSTAAQAFRTDSPETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLVVLDPSKEVAPMIIKHRSQAGRDVHILD
PRNRDTGFNALDWIGRHGGTKEEDIAAVASWIMSDNGGARGVRDDFFRASALQLLTALIADVCLSGHTLK
EHQTLRQVRKNLAEPEPKLRERLQRIHGNSKSEFVKENVASFVNMTPETFSGVYANAVKETHWLSYLNYA
ALVSGSSFSTDELARGDLDVFINIDLKTLEAHSGLARVIIGSFLNAIYNHDGDLKGRALFLLDEVARLGY
MRILETARDAGRKYGISLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VEIDQVSRSFQSKGTSRTRSKQLAARPLIQPHEVLRMRPDEQIVFTAGNPPLRCGRAIWFRRDDMRMRVV
ENRFHALAGGAKERLNSSHPAKIDEITTGVRSVGEEWNEIGQEPHDPFQEES
MTANKIALVGLPITLMILVTIGMTGSEQWLSAFGKTEAGRRMLGRAGIALPYIASATVGMIVLFASAGST
NIKLAGWSVVGGSSVTILIAALRETVRLVALLDQLPTARSALFYADPATMIGAAAALISAAFALRVSLVG
NAAFATAEPKRILGRRALHGEADWMKMTQAGKLFPDAGGIVIGERYRVDKDSTAAQAFRTDSPETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLVVLDPSKEVAPMIIKHRSQAGRDVHILD
PRNRDTGFNALDWIGRHGGTKEEDIAAVASWIMSDNGGARGVRDDFFRASALQLLTALIADVCLSGHTLK
EHQTLRQVRKNLAEPEPKLRERLQRIHGNSKSEFVKENVASFVNMTPETFSGVYANAVKETHWLSYLNYA
ALVSGSSFSTDELARGDLDVFINIDLKTLEAHSGLARVIIGSFLNAIYNHDGDLKGRALFLLDEVARLGY
MRILETARDAGRKYGISLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISKRCGMTT
VEIDQVSRSFQSKGTSRTRSKQLAARPLIQPHEVLRMRPDEQIVFTAGNPPLRCGRAIWFRRDDMRMRVV
ENRFHALAGGAKERLNSSHPAKIDEITTGVRSVGEEWNEIGQEPHDPFQEES
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1559 | GenBank | WP_130719428 |
| Name | t4cp2_ELH90_RS35150_pSM135B_Rh04 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 91370.94 Da Isoelectric Point: 5.2674
>WP_130719428.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVSLRVFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDLERNEISRQINAILSRLG
SGWMIQVEAVRLATVDYAAAGKSDFPDRVTRAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSTLSK
YIYSDEGSRKQSYADTVLFIFRNAIREVEQYLANVISIRRMMTRETTERGGARVARYDELLQFIRFCVTG
DNYPIRLPDVPMYLDWIATAELQHGLAPKVENRFLGVVAVDGLPSESWPGILNNLDLMPLSYRWSSRFIF
LDAEEAKVRLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMSMVAETEHAIAEASSQLVAYGYYTPVVVL
FENDPERLKEQMEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNVREPLINTRNLSDLIPLNSVWS
GNPQAPCPFYPPDSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYEDAQIFA
FDKGGSMMPLTLAVGGDHYEVGGDEGTGLSFCPLSDLSSEGDRAWACEWIESLVAMQGITITPDHRNAIS
RQIGLMAGAPGRSISDFVSGVQIREIKDALNHYTIDGPMGHLLDAETDGLALGVFQTFEIEQVMNTGERN
LVPVLTYLFRRIEKRLTGAPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREPGTRDFYERIGFNDRQIEIVATAIPKREYYVASPEGRRLFDMALGPLTL
SFVGASGKEDLKRVRALSEEYGPEWPIQWLQTRGIKDAASLLEDA
MVSLRVFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDLERNEISRQINAILSRLG
SGWMIQVEAVRLATVDYAAAGKSDFPDRVTRAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSTLSK
YIYSDEGSRKQSYADTVLFIFRNAIREVEQYLANVISIRRMMTRETTERGGARVARYDELLQFIRFCVTG
DNYPIRLPDVPMYLDWIATAELQHGLAPKVENRFLGVVAVDGLPSESWPGILNNLDLMPLSYRWSSRFIF
LDAEEAKVRLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMSMVAETEHAIAEASSQLVAYGYYTPVVVL
FENDPERLKEQMEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNVREPLINTRNLSDLIPLNSVWS
GNPQAPCPFYPPDSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYEDAQIFA
FDKGGSMMPLTLAVGGDHYEVGGDEGTGLSFCPLSDLSSEGDRAWACEWIESLVAMQGITITPDHRNAIS
RQIGLMAGAPGRSISDFVSGVQIREIKDALNHYTIDGPMGHLLDAETDGLALGVFQTFEIEQVMNTGERN
LVPVLTYLFRRIEKRLTGAPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREPGTRDFYERIGFNDRQIEIVATAIPKREYYVASPEGRRLFDMALGPLTL
SFVGASGKEDLKRVRALSEEYGPEWPIQWLQTRGIKDAASLLEDA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 369288..378765
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH90_RS35095 (ELH90_35095) | 364745..366139 | - | 1395 | WP_130719417 | amidohydrolase family protein | - |
| ELH90_RS40105 (ELH90_35100) | 366129..366680 | - | 552 | WP_342635959 | response regulator transcription factor | - |
| ELH90_RS35105 (ELH90_35105) | 367454..368191 | + | 738 | WP_130719419 | response regulator transcription factor | - |
| ELH90_RS35110 (ELH90_35110) | 368579..369262 | + | 684 | WP_245499002 | autoinducer binding domain-containing protein | - |
| ELH90_RS35115 (ELH90_35115) | 369288..370583 | - | 1296 | WP_130719421 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH90_RS35120 (ELH90_35120) | 370596..371024 | - | 429 | WP_130719422 | conjugal transfer protein TrbH | - |
| ELH90_RS35125 (ELH90_35125) | 371027..371848 | - | 822 | WP_130719423 | P-type conjugative transfer protein TrbG | virB9 |
| ELH90_RS35130 (ELH90_35130) | 371865..372527 | - | 663 | WP_130719424 | conjugal transfer protein TrbF | virB8 |
| ELH90_RS35135 (ELH90_35135) | 372551..373729 | - | 1179 | WP_130719425 | P-type conjugative transfer protein TrbL | virB6 |
| ELH90_RS35145 (ELH90_35145) | 373911..374711 | - | 801 | WP_130719427 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH90_RS35150 (ELH90_35150) | 374683..377130 | - | 2448 | WP_130719428 | conjugal transfer protein TrbE | virb4 |
| ELH90_RS35155 (ELH90_35155) | 377141..377437 | - | 297 | WP_130719441 | conjugal transfer protein TrbD | virB3 |
| ELH90_RS35160 (ELH90_35160) | 377430..377810 | - | 381 | WP_130719429 | TrbC/VirB2 family protein | virB2 |
| ELH90_RS35165 (ELH90_35165) | 377800..378765 | - | 966 | WP_130719430 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH90_RS35170 (ELH90_35170) | 378779..379408 | - | 630 | WP_130719431 | acyl-homoserine-lactone synthase TraI | - |
| ELH90_RS35175 (ELH90_35175) | 379772..380989 | + | 1218 | WP_018247073 | plasmid partitioning protein RepA | - |
| ELH90_RS35180 (ELH90_35180) | 380974..382014 | + | 1041 | WP_018448901 | plasmid partitioning protein RepB | - |
| ELH90_RS35185 (ELH90_35185) | 382254..383519 | + | 1266 | WP_130719432 | plasmid replication protein RepC | - |
Host bacterium
| ID | 3026 | GenBank | NZ_SIOP01000004 |
| Plasmid name | pSM135B_Rh04 | Incompatibility group | - |
| Plasmid size | 388771 bp | Coordinate of oriT [Strand] | 349132..349167 [+] |
| Host baterium | Rhizobium leguminosarum strain SM135B |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | noeL |
| Anti-CRISPR | AcrIIA7 |