Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102577 |
| Name | oriT_pSM126A_Rh08 |
| Organism | Rhizobium leguminosarum strain SM126A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIOA01000005 (10760..10793 [+], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pSM126A_Rh08
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file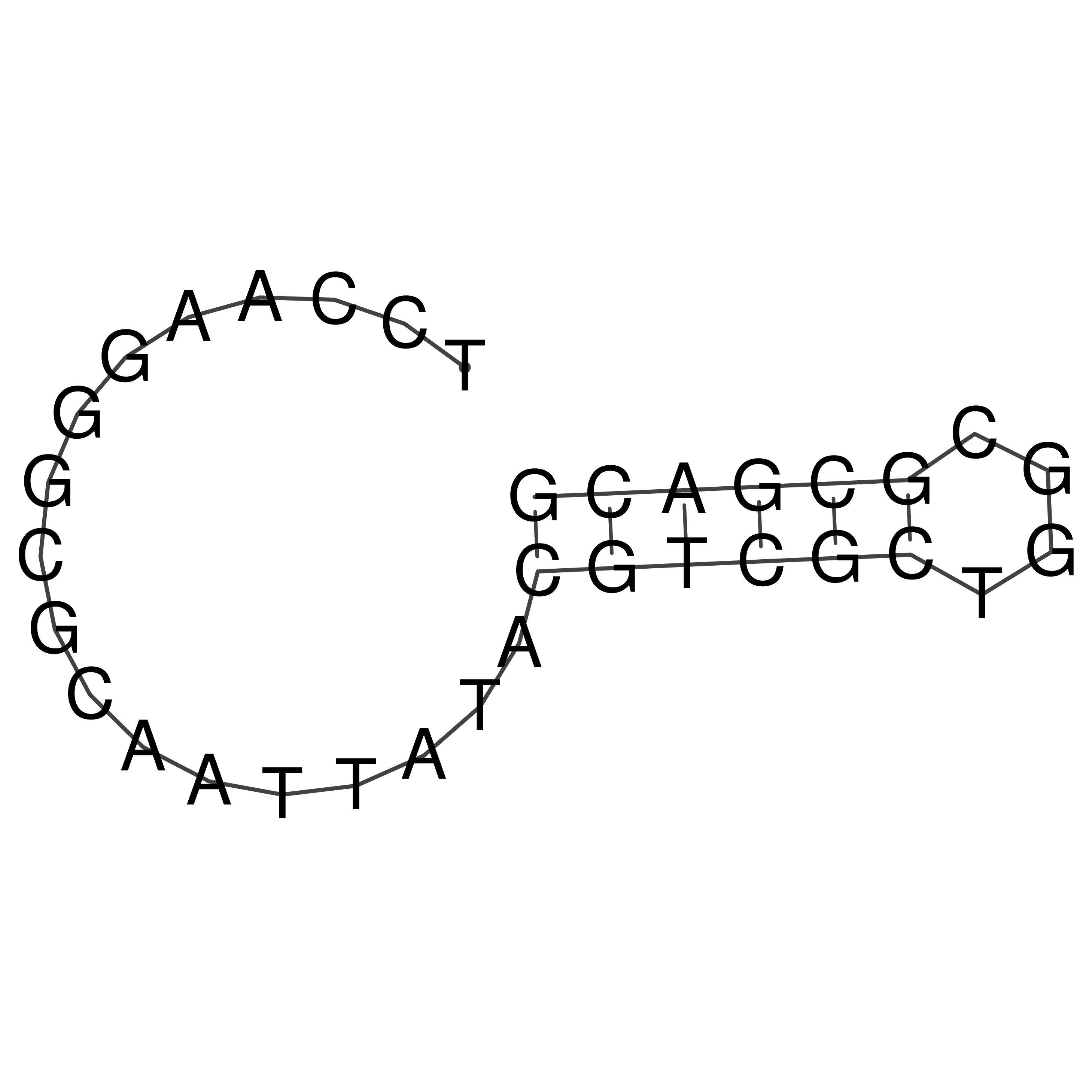
Relaxase
| ID | 1925 | GenBank | WP_130657460 |
| Name | traA_ELH75_RS31615_pSM126A_Rh08 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122987.83 Da Isoelectric Point: 9.6219
>WP_130657460.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKVEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSSAAKQTLERVRDVIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTDGQKAEVQSTWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKPIRNDAGKFVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKVEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQGPEALRLERERINFATGSRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLEATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWEIRWNQSRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDAALDLARGNVGKA
VDAYRGYGHVRGLDLKAQAVESLIADWNHGYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLGKVVEASRNRIVAEVGEGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYD
KASFYGQALRFAEARGFHLVNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSFPDTI
KEAKPMVSGITTFPQTLDQAVEDKLAADPGLKKQWEDVSTRFQLVYATPEAAFKAIDVDTMLKDQSVAQS
TLARIAGEPESFGAIKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAELKHETEERAVRLKV
SVDIPALSSAAKQTLERVRDVIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTDGQKAEVQSTWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1552 | GenBank | WP_130759824 |
| Name | traG_ELH75_RS31600_pSM126A_Rh08 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69558.74 Da Isoelectric Point: 8.9360
>WP_130759824.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSDHTDE
KDQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQLNGRALFLLDEVARLGY
MRILETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
MTINRIALAVMPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRTGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTETLNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRIDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSDHTDE
KDQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGKTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQLNGRALFLLDEVARLGY
MRILETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1553 | GenBank | WP_065276635 |
| Name | t4cp2_ELH75_RS31715_pSM126A_Rh08 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91588.38 Da Isoelectric Point: 5.9977
>WP_065276635.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNALRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNALRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRTLKSEHGHDWPIHWLETRGVHDAASLFK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8183..32757
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH75_RS37450 | 3565..3738 | + | 174 | WP_165403408 | hypothetical protein | - |
| ELH75_RS31580 (ELH75_31575) | 3806..4114 | - | 309 | WP_130657465 | WGR domain-containing protein | - |
| ELH75_RS31585 (ELH75_31580) | 4465..5958 | - | 1494 | WP_130657464 | group II intron reverse transcriptase/maturase | - |
| ELH75_RS31590 (ELH75_31585) | 6740..7054 | + | 315 | WP_130657463 | hypothetical protein | - |
| ELH75_RS31595 (ELH75_31590) | 7297..7800 | - | 504 | WP_197726741 | thermonuclease family protein | - |
| ELH75_RS31600 (ELH75_31595) | 8183..10117 | - | 1935 | WP_130759824 | Ti-type conjugative transfer system protein TraG | virb4 |
| ELH75_RS31605 (ELH75_31600) | 10104..10319 | - | 216 | WP_027668129 | type IV conjugative transfer system coupling protein TraD | - |
| ELH75_RS31610 (ELH75_31605) | 10324..10617 | - | 294 | WP_011654562 | conjugal transfer protein TraC | - |
| ELH75_RS31615 (ELH75_31610) | 10873..14199 | + | 3327 | WP_130657460 | Ti-type conjugative transfer relaxase TraA | - |
| ELH75_RS31620 (ELH75_31615) | 14196..14759 | + | 564 | WP_130657459 | conjugative transfer signal peptidase TraF | - |
| ELH75_RS31625 (ELH75_31620) | 14749..15912 | + | 1164 | WP_130657458 | conjugal transfer protein TraB | - |
| ELH75_RS31630 (ELH75_31625) | 15929..16546 | + | 618 | WP_130657457 | TraH family protein | virB1 |
| ELH75_RS31640 (ELH75_31635) | 18371..18718 | - | 348 | WP_018244911 | IS66 family insertion sequence element accessory protein TnpB | - |
| ELH75_RS31645 (ELH75_31640) | 18715..19125 | - | 411 | WP_018244910 | transposase | - |
| ELH75_RS31650 (ELH75_31645) | 19336..20204 | - | 869 | Protein_19 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| ELH75_RS31655 (ELH75_31650) | 20197..20814 | - | 618 | WP_130654291 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| ELH75_RS38010 (ELH75_31655) | 20905..21336 | - | 432 | Protein_21 | hypothetical protein | - |
| ELH75_RS31665 (ELH75_31660) | 21439..21681 | - | 243 | WP_130654290 | helix-turn-helix transcriptional regulator | - |
| ELH75_RS31670 (ELH75_31665) | 21904..22227 | + | 324 | WP_130657456 | transcriptional repressor TraM | - |
| ELH75_RS31675 (ELH75_31670) | 22231..22935 | - | 705 | WP_130759825 | autoinducer binding domain-containing protein | - |
| ELH75_RS31680 (ELH75_31675) | 23246..24544 | - | 1299 | WP_130660093 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH75_RS31685 (ELH75_31680) | 24556..24990 | - | 435 | WP_130657454 | conjugal transfer protein TrbH | - |
| ELH75_RS31690 (ELH75_31685) | 24994..25806 | - | 813 | WP_065276640 | P-type conjugative transfer protein TrbG | virB9 |
| ELH75_RS31695 (ELH75_31690) | 25824..26486 | - | 663 | WP_065276639 | conjugal transfer protein TrbF | virB8 |
| ELH75_RS31700 (ELH75_31695) | 26507..27688 | - | 1182 | WP_065276638 | P-type conjugative transfer protein TrbL | virB6 |
| ELH75_RS31705 (ELH75_31700) | 27682..27882 | - | 201 | WP_065276637 | entry exclusion protein TrbK | - |
| ELH75_RS31710 (ELH75_31705) | 27879..28676 | - | 798 | WP_130657453 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH75_RS31715 (ELH75_31710) | 28654..31104 | - | 2451 | WP_065276635 | conjugal transfer protein TrbE | virb4 |
| ELH75_RS31720 (ELH75_31715) | 31115..31414 | - | 300 | WP_130654279 | conjugal transfer protein TrbD | virB3 |
| ELH75_RS31725 (ELH75_31720) | 31407..31790 | - | 384 | WP_065276633 | TrbC/VirB2 family protein | virB2 |
| ELH75_RS31730 (ELH75_31725) | 31780..32757 | - | 978 | WP_065276632 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH75_RS31735 (ELH75_31730) | 32754..33392 | - | 639 | WP_130660092 | acyl-homoserine-lactone synthase | - |
| ELH75_RS31740 (ELH75_31735) | 33769..34992 | + | 1224 | WP_065276670 | plasmid partitioning protein RepA | - |
| ELH75_RS31745 (ELH75_31740) | 34989..36026 | + | 1038 | WP_130657451 | plasmid partitioning protein RepB | - |
| ELH75_RS31750 (ELH75_31745) | 36216..37430 | + | 1215 | WP_065276629 | plasmid replication protein RepC | - |
Host bacterium
| ID | 3021 | GenBank | NZ_SIOA01000005 |
| Plasmid name | pSM126A_Rh08 | Incompatibility group | - |
| Plasmid size | 41634 bp | Coordinate of oriT [Strand] | 10760..10793 [+] |
| Host baterium | Rhizobium leguminosarum strain SM126A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |