Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102575 |
| Name | oriT_pSM138B_Rh13 |
| Organism | Rhizobium leguminosarum strain SM138B |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIOU01000007 (131510..131543 [+], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pSM138B_Rh13
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file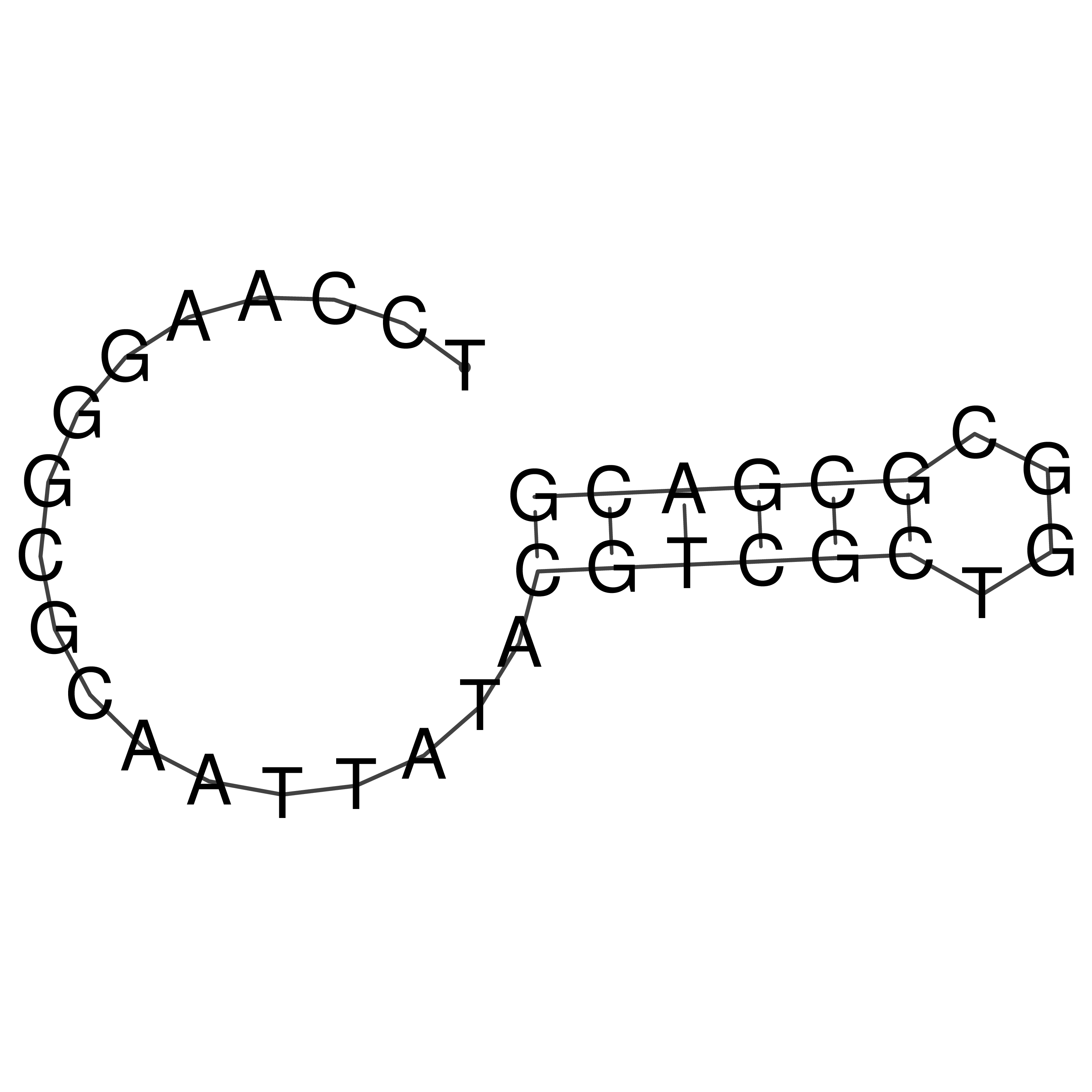
Relaxase
| ID | 1923 | GenBank | WP_130747837 |
| Name | traA_ELH95_RS31510_pSM138B_Rh13 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122627.81 Da Isoelectric Point: 9.9195
>WP_130747837.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEAFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLAEDGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSPPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDNVALFQSLMVRILQSPEALRLERERINFAIGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLQATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIGSRTLSSWELRWNQGRNQLDHKTVFVLDEAGMVSSRQM
ALLVETVTKAGAKLVLVGDPEQLQPIAAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRKLNDMARAKLVERSVIADGSAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEATPGRVVAELGEGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGFHLVNVARTIADDRLQWAVRQKQKLADLGARLAAIAAALGLVRGSNKHSIPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYANPEAAFKAINVDTMLKDQSVAQS
TLARVAGEPESFGALKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLDYALADKMVKAELEGFAKAVSERFGERTFLSLAAKDTSG
KTFETVTAGMTAGQKAEVQSAWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEAFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLAEDGFGSKKVAVLGPDGKPIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSPPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDNVALFQSLMVRILQSPEALRLERERINFAIGIRTPAKYTTREM
IRLEAEMANRAIWLSGRASHGVREAVLQATFARHSRLSDEQRTAIEHVAGATRIAAVIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIGSRTLSSWELRWNQGRNQLDHKTVFVLDEAGMVSSRQM
ALLVETVTKAGAKLVLVGDPEQLQPIAAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRKLNDMARAKLVERSVIADGSAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEATPGRVVAELGEGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGFHLVNVARTIADDRLQWAVRQKQKLADLGARLAAIAAALGLVRGSNKHSIPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYANPEAAFKAINVDTMLKDQSVAQS
TLARVAGEPESFGALKGKTGLLASRGDKQDREKAIANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLDYALADKMVKAELEGFAKAVSERFGERTFLSLAAKDTSG
KTFETVTAGMTAGQKAEVQSAWNSMRTVQQLAAYERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1548 | GenBank | WP_130747834 |
| Name | traG_ELH95_RS31495_pSM138B_Rh13 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69379.45 Da Isoelectric Point: 9.3498
>WP_130747834.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYVAGAVIGVLLLFASAGSI
NIKQAGWGVVAGSSGTILVAAVRETVRLSSFMKTVPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAQLFPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGGAGRDVFILD
PRKPDVGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGSRGVRDDFFRASALQLLTALIADVCLSGHTDE
KEQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKASVG
PNRFHMIGDTPKPA
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYVAGAVIGVLLLFASAGSI
NIKQAGWGVVAGSSGTILVAAVRETVRLSSFMKTVPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAQLFPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGGAGRDVFILD
PRKPDVGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGSRGVRDDFFRASALQLLTALIADVCLSGHTDE
KEQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKASVG
PNRFHMIGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1549 | GenBank | WP_130747842 |
| Name | t4cp2_ELH95_RS31605_pSM138B_Rh13 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91606.33 Da Isoelectric Point: 5.7770
>WP_130747842.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTLAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVIAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYAPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSSLLPLTLAADGDHYEIGGDNAEEGRALAFCPLSDLKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPALTYLFRRIEKRLEGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREFGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSEHGRHWPIHWLETRGVHDAASLFK
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTLAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVIAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYAPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGSSLLPLTLAADGDHYEIGGDNAEEGRALAFCPLSDLKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPALTYLFRRIEKRLEGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREFGTREFYERIGFNERQIEIVATAMPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSEHGRHWPIHWLETRGVHDAASLFK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 136682..152411
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH95_RS31515 (ELH95_31520) | 135003..135512 | + | 510 | WP_245487465 | conjugative transfer signal peptidase TraF | - |
| ELH95_RS31520 (ELH95_31525) | 135502..136665 | + | 1164 | WP_130679582 | conjugal transfer protein TraB | - |
| ELH95_RS31525 (ELH95_31530) | 136682..137302 | + | 621 | WP_130679581 | TraH family protein | virB1 |
| ELH95_RS31530 (ELH95_31535) | 137483..137932 | + | 450 | WP_130679913 | GNAT family N-acetyltransferase | - |
| ELH95_RS31535 (ELH95_31540) | 138090..138917 | - | 828 | WP_245487467 | aminoglycoside phosphotransferase family protein | - |
| ELH95_RS31540 (ELH95_31545) | 139208..139582 | - | 375 | WP_087003573 | DUF5615 family PIN-like protein | - |
| ELH95_RS31545 (ELH95_31550) | 139579..140211 | - | 633 | WP_130679580 | DUF433 domain-containing protein | - |
| ELH95_RS31550 (ELH95_31555) | 140332..140580 | - | 249 | WP_130679579 | helix-turn-helix transcriptional regulator | - |
| ELH95_RS31555 (ELH95_31560) | 140872..141162 | + | 291 | WP_130679578 | transcriptional repressor TraM | - |
| ELH95_RS31560 (ELH95_31565) | 141166..141870 | - | 705 | WP_130679577 | autoinducer binding domain-containing protein | - |
| ELH95_RS31565 (ELH95_31570) | 141976..142716 | - | 741 | WP_130747838 | LuxR family transcriptional regulator | - |
| ELH95_RS31570 (ELH95_31575) | 142900..144195 | - | 1296 | WP_130747839 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH95_RS31575 (ELH95_31580) | 144207..144647 | - | 441 | WP_130747840 | conjugal transfer protein TrbH | - |
| ELH95_RS31580 (ELH95_31585) | 144651..145463 | - | 813 | WP_130747841 | P-type conjugative transfer protein TrbG | virB9 |
| ELH95_RS31585 (ELH95_31590) | 145481..146143 | - | 663 | WP_130679572 | conjugal transfer protein TrbF | virB8 |
| ELH95_RS31590 (ELH95_31595) | 146164..147345 | - | 1182 | WP_130679571 | P-type conjugative transfer protein TrbL | virB6 |
| ELH95_RS31595 (ELH95_31600) | 147339..147539 | - | 201 | WP_130679570 | entry exclusion protein TrbK | - |
| ELH95_RS31600 (ELH95_31605) | 147536..148333 | - | 798 | WP_130679569 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH95_RS31605 (ELH95_31610) | 148311..150761 | - | 2451 | WP_130747842 | conjugal transfer protein TrbE | virb4 |
| ELH95_RS31610 (ELH95_31615) | 150772..151071 | - | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELH95_RS31615 (ELH95_31620) | 151064..151444 | - | 381 | WP_130747843 | TrbC/VirB2 family protein | virB2 |
| ELH95_RS31620 (ELH95_31625) | 151434..152411 | - | 978 | WP_130747844 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH95_RS31625 (ELH95_31630) | 152422..153042 | - | 621 | WP_130747845 | acyl-homoserine-lactone synthase | - |
| ELH95_RS31630 (ELH95_31635) | 153422..154618 | + | 1197 | WP_130747846 | plasmid partitioning protein RepA | - |
| ELH95_RS31635 (ELH95_31640) | 154742..155701 | + | 960 | WP_130747847 | plasmid partitioning protein RepB | - |
| ELH95_RS31640 (ELH95_31645) | 155874..157091 | + | 1218 | WP_130747848 | plasmid replication protein RepC | - |
Host bacterium
| ID | 3019 | GenBank | NZ_SIOU01000007 |
| Plasmid name | pSM138B_Rh13 | Incompatibility group | - |
| Plasmid size | 183277 bp | Coordinate of oriT [Strand] | 131510..131543 [+] |
| Host baterium | Rhizobium leguminosarum strain SM138B |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |