Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102557 |
| Name | oriT_pSM27_Rh04 |
| Organism | Rhizobium leguminosarum strain SM27 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIKL01000005 (57676..57716 [-], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pSM27_Rh04
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file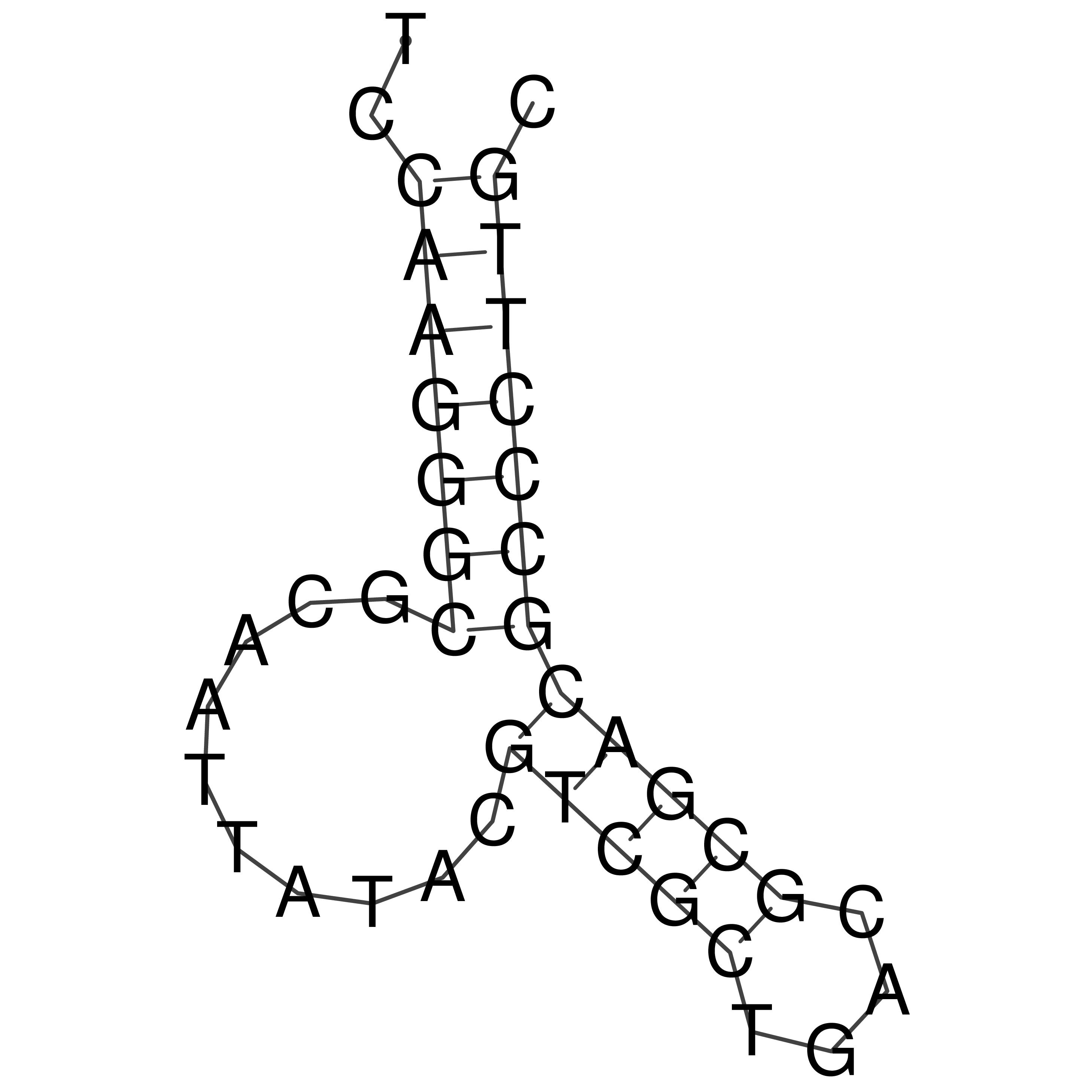
Relaxase
| ID | 1908 | GenBank | WP_130801322 |
| Name | traA_ELG82_RS32300_pSM27_Rh04 |
UniProt ID | _ |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 122944.16 Da Isoelectric Point: 10.2420
>WP_130801322.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKRGLLHEEFVIPVWAPTWLRAMIADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELSAEQNVALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKIAVASPDGNPMRNGDGKIVYELWAGSADDFNAIRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIDRKAEQANARTSAWSPKLERVELQEDRRAENARRMQRRPE
IVLDLITREKSVFDERDVAKILHRYIDDAALFQNLMVRILQSPEALRLQRDRIDFATATRTPAKYTTREM
IRLEAEMVNRAIWLSGRASHGVREAVLEVTFARHARLSDEQRTAIERVAGGERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKQAGIQSRTLSSWELRWNQGRNQIDARTVFVLDEAGMVSSRQM
AHFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRTQWMRDASLDLARGNVGKA
VDDYRAHGHVRGLDLKAQAVENLIADWNHDYDPSKSSLILAHLRRDVRMLNDMARAKLVDRGIVVDGFAF
TTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRLVVALGEGDHRRLVTIEQRFYNKLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILARRKAKETTLDYE
KSSLYGQALRFAEARGLHLMNVARTVARDRLEWTVRQKQKLADLGARLAAIGVKLGRAASAKTSPNPNAK
ESKPMVSGISTFPKSIDQSVEDKLAADPGLKKQWEDVSTRFHLVYAQPESAFKAVNVEAMLKDQAIAKTT
LAKLAGEPERFGALRGKTGVFANRGDKQDRERAIANVPALARNLERYLRQRAEVERKHEREERAVRRQVS
VDIPALSASAKQTLERVRDAIDRNDLSAGLEYALADKMVKAELDGFAKAVSERFGERTFLGIASKDPSGV
TFKSVTAGMNPAQKAEVQIAWNSMRSIQQLSADEKTSEALKQAETLRQARSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTRKRGLLHEEFVIPVWAPTWLRAMIADR
SVSGASEAFWNKVEAFEKRSDAQLAKDVTIALPIELSAEQNVALVRDFVERHITSKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKIAVASPDGNPMRNGDGKIVYELWAGSADDFNAIRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIELTPTIHLGVGTKAIDRKAEQANARTSAWSPKLERVELQEDRRAENARRMQRRPE
IVLDLITREKSVFDERDVAKILHRYIDDAALFQNLMVRILQSPEALRLQRDRIDFATATRTPAKYTTREM
IRLEAEMVNRAIWLSGRASHGVREAVLEVTFARHARLSDEQRTAIERVAGGERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKQAGIQSRTLSSWELRWNQGRNQIDARTVFVLDEAGMVSSRQM
AHFVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRTQWMRDASLDLARGNVGKA
VDDYRAHGHVRGLDLKAQAVENLIADWNHDYDPSKSSLILAHLRRDVRMLNDMARAKLVDRGIVVDGFAF
TTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRLVVALGEGDHRRLVTIEQRFYNKLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILARRKAKETTLDYE
KSSLYGQALRFAEARGLHLMNVARTVARDRLEWTVRQKQKLADLGARLAAIGVKLGRAASAKTSPNPNAK
ESKPMVSGISTFPKSIDQSVEDKLAADPGLKKQWEDVSTRFHLVYAQPESAFKAVNVEAMLKDQAIAKTT
LAKLAGEPERFGALRGKTGVFANRGDKQDRERAIANVPALARNLERYLRQRAEVERKHEREERAVRRQVS
VDIPALSASAKQTLERVRDAIDRNDLSAGLEYALADKMVKAELDGFAKAVSERFGERTFLGIASKDPSGV
TFKSVTAGMNPAQKAEVQIAWNSMRSIQQLSADEKTSEALKQAETLRQARSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1524 | GenBank | WP_130801259 |
| Name | t4cp2_ELG82_RS32065_pSM27_Rh04 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 91388.01 Da Isoelectric Point: 6.1930
>WP_130801259.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVSLRAFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDFERNEVSRQINAILSRLG
SGWMIQVEAVRLATVDYASAGKSHFPDRVTLAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSRLSK
YIYSDEDSRKQSYADTVLFIFRNAIREVEQYLANIISIRRMLTRETAERGGVRVAHYDELLQFIRFCVTG
ENHPVRVPDIPMYLDWIATAELRHGLAPKVENRFLGVVAIDGLPSESWPGILNSLDAMPLGYRWSSRFIF
LDAEEARVRLERTRKKWLQKVRPFFDQLFQTQSRSVDQDAISMVAETEDAIAQASSQLVAYGYYTPVVVL
FDNDAERLNEKTEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTRNLSDLIPLNSVWS
GNPQAPCPFYPPGSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGRSMLPLTLACGGDHYEIGGDEDAGLSFCPLSELSSDGDRAWASEWIESLVAMQGITITPDHRNAIS
RQIGLMDGAPGRSISDFVSGIQMREIKDALHHYTVDGPMGQLLDAETDGLALGLFQTFEIEQVMNMGERN
LVPVLTYLFRRIEKRLTGAPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREAGTRDFYERIGFNERQIEIVATAIPKREYYVASPEGRRLFDMALGPLAL
SFVGASSKDDLKHIRALHGEHGPEWPLHWLHKRGIKDAASLLKDA
MVSLRAFRHTAPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESSTDFERNEVSRQINAILSRLG
SGWMIQVEAVRLATVDYASAGKSHFPDRVTLAIDEERRAHFEREQGHFESRHAIIATYRPTEQRRSRLSK
YIYSDEDSRKQSYADTVLFIFRNAIREVEQYLANIISIRRMLTRETAERGGVRVAHYDELLQFIRFCVTG
ENHPVRVPDIPMYLDWIATAELRHGLAPKVENRFLGVVAIDGLPSESWPGILNSLDAMPLGYRWSSRFIF
LDAEEARVRLERTRKKWLQKVRPFFDQLFQTQSRSVDQDAISMVAETEDAIAQASSQLVAYGYYTPVVVL
FDNDAERLNEKTEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTRNLSDLIPLNSVWS
GNPQAPCPFYPPGSPPLMQVATGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGRSMLPLTLACGGDHYEIGGDEDAGLSFCPLSELSSDGDRAWASEWIESLVAMQGITITPDHRNAIS
RQIGLMDGAPGRSISDFVSGIQMREIKDALHHYTVDGPMGQLLDAETDGLALGLFQTFEIEQVMNMGERN
LVPVLTYLFRRIEKRLTGAPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAERSGI
IDVLKESCPTKICLPNGAAREAGTRDFYERIGFNERQIEIVATAIPKREYYVASPEGRRLFDMALGPLAL
SFVGASSKDDLKHIRALHGEHGPEWPLHWLHKRGIKDAASLLKDA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1525 | GenBank | WP_130801328 |
| Name | traG_ELG82_RS32315_pSM27_Rh04 |
UniProt ID | _ |
| Length | 655 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 655 a.a. Molecular weight: 71008.84 Da Isoelectric Point: 9.0366
>WP_130801328.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTANKIALVSVPIALMILVTIGMTGIERWFSEFGKTETARLTLGRTGIALPYIASAATGIIFLFASAGST
NIKLAGWGVVGGSSATIVIAALRETVRLAAIFDHFPAVRSALSYADPATMIGAGAALTSAGFALRVARLG
NVAFARSEPRRIRGRRALHGEAEWMKSTEAGKLFPDTGGIVIGERYRVDQDMTAKQSFRPDSVETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLVLLDPSNEVAPMVIGHRTNAGRDVHILD
PRTCDTGFNALDWIGLHGRTKEEDIAAVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGRTEE
GDQTLRQVRKNLSEPEPKLRARLQSIYDNSRSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSESTFSTNDISAGNADVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGALKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRACVG
ENRFHSPASGDESAMDPETPMKLRG
MTANKIALVSVPIALMILVTIGMTGIERWFSEFGKTETARLTLGRTGIALPYIASAATGIIFLFASAGST
NIKLAGWGVVGGSSATIVIAALRETVRLAAIFDHFPAVRSALSYADPATMIGAGAALTSAGFALRVARLG
NVAFARSEPRRIRGRRALHGEAEWMKSTEAGKLFPDTGGIVIGERYRVDQDMTAKQSFRPDSVETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLVLLDPSNEVAPMVIGHRTNAGRDVHILD
PRTCDTGFNALDWIGLHGRTKEEDIAAVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGRTEE
GDQTLRQVRKNLSEPEPKLRARLQSIYDNSRSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSESTFSTNDISAGNADVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGALKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMRACVG
ENRFHSPASGDESAMDPETPMKLRG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 12763..22142
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELG82_RS32030 (ELG82_32035) | 8012..9277 | - | 1266 | WP_130801245 | plasmid replication protein RepC | - |
| ELG82_RS32035 (ELG82_32040) | 9516..10556 | - | 1041 | WP_130801247 | plasmid partitioning protein RepB | - |
| ELG82_RS32040 (ELG82_32045) | 10541..11758 | - | 1218 | WP_130801249 | plasmid partitioning protein RepA | - |
| ELG82_RS32045 (ELG82_32050) | 12123..12749 | + | 627 | WP_130801251 | acyl-homoserine-lactone synthase TraI | - |
| ELG82_RS32050 (ELG82_32055) | 12763..13728 | + | 966 | WP_130801253 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELG82_RS32055 (ELG82_32060) | 13718..14098 | + | 381 | WP_130801255 | TrbC/VirB2 family protein | virB2 |
| ELG82_RS32060 (ELG82_32065) | 14088..14387 | + | 300 | WP_130801257 | conjugal transfer protein TrbD | virB3 |
| ELG82_RS32065 (ELG82_32070) | 14397..16844 | + | 2448 | WP_130801259 | conjugal transfer protein TrbE | virb4 |
| ELG82_RS32070 (ELG82_32075) | 16816..17616 | + | 801 | WP_130801261 | P-type conjugative transfer protein TrbJ | virB5 |
| ELG82_RS32075 (ELG82_32080) | 17748..18896 | + | 1149 | WP_130801330 | P-type conjugative transfer protein TrbL | virB6 |
| ELG82_RS32080 (ELG82_32085) | 18916..19578 | + | 663 | WP_130801263 | conjugal transfer protein TrbF | virB8 |
| ELG82_RS32085 (ELG82_32090) | 19595..20416 | + | 822 | WP_130801265 | P-type conjugative transfer protein TrbG | virB9 |
| ELG82_RS32090 (ELG82_32095) | 20419..20847 | + | 429 | WP_130801267 | conjugal transfer protein TrbH | - |
| ELG82_RS32095 (ELG82_32100) | 20862..22142 | + | 1281 | WP_130801269 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELG82_RS32100 (ELG82_32105) | 22168..22878 | - | 711 | WP_130801270 | autoinducer binding domain-containing protein | - |
| ELG82_RS32105 (ELG82_32110) | 23137..23343 | + | 207 | Protein_23 | IS6 family transposase | - |
| ELG82_RS32110 (ELG82_32115) | 24031..25992 | + | 1962 | WP_130801271 | elongation factor G | - |
| ELG82_RS32115 (ELG82_32120) | 26406..26825 | + | 420 | WP_245505619 | lysophospholipase | - |
Host bacterium
| ID | 3001 | GenBank | NZ_SIKL01000005 |
| Plasmid name | pSM27_Rh04 | Incompatibility group | - |
| Plasmid size | 62563 bp | Coordinate of oriT [Strand] | 57676..57716 [-] |
| Host baterium | Rhizobium leguminosarum strain SM27 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |