Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102553 |
| Name | oriT_pSM16_Rh07 |
| Organism | Rhizobium leguminosarum strain SM16 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIKC01000007 (195075..195115 [-], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pSM16_Rh07
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file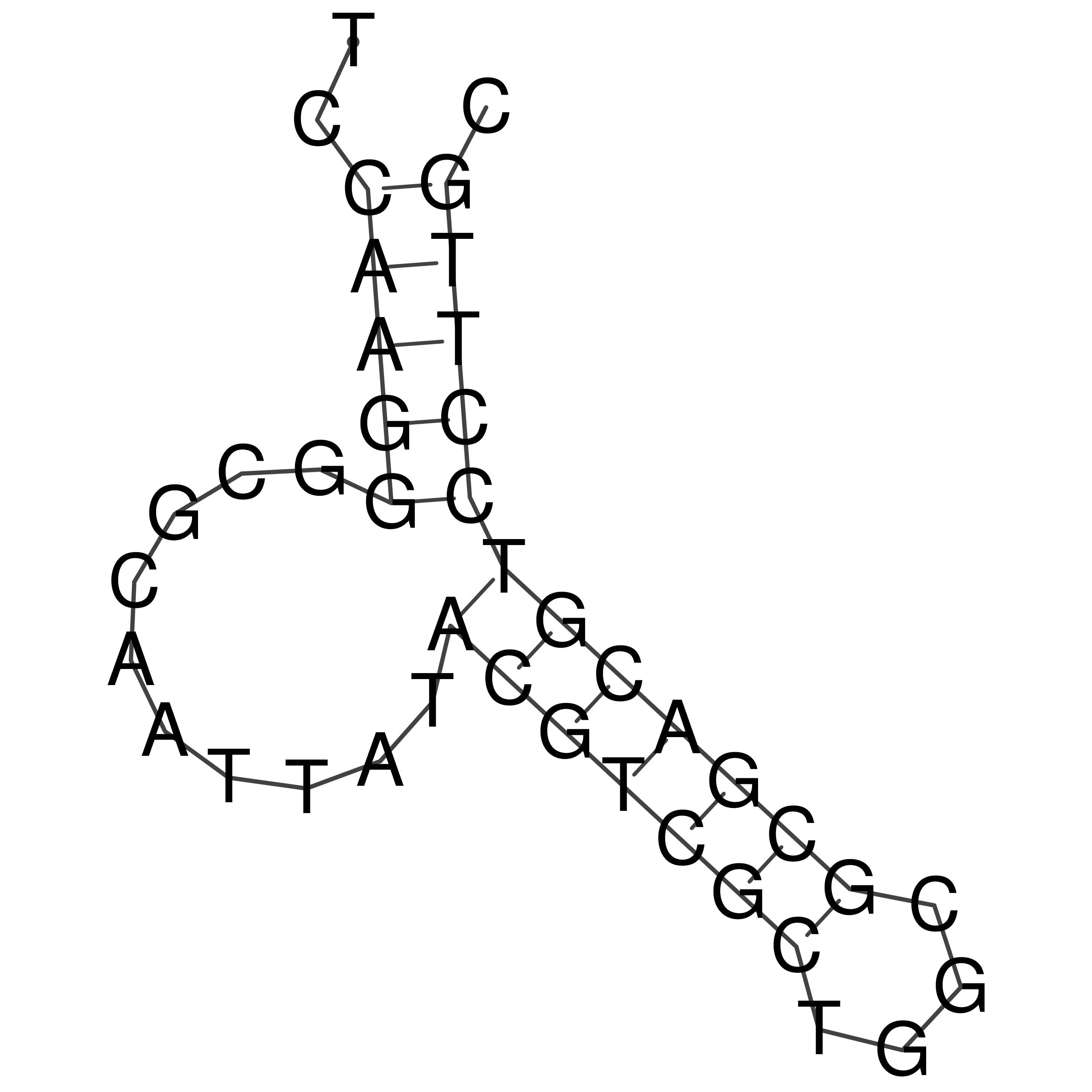
Relaxase
| ID | 1904 | GenBank | WP_130784879 |
| Name | traA_ELG73_RS33500_pSM16_Rh07 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122892.96 Da Isoelectric Point: 9.7033
>WP_130784879.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVCSMIADR
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKIALLGPDGKPVRNDAGKLVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKTQASAPKLERIELQGERRSENARRIERRPE
IVLDLITREKSVFDERDVAKVLYRYIDDAALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTRDM
VRLEAEMANRAIWLSGRASHGVREEVLQTTFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLESAPGRIVAEVGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLVNVARTIAHDRIHWAVRQKQNLADLGARLAAISTALGLARGSNKHSIPNTI
KGAKPMVSGITTFPKSLDHAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAVFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGVLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVCSMIADR
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKIALLGPDGKPVRNDAGKLVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKTQASAPKLERIELQGERRSENARRIERRPE
IVLDLITREKSVFDERDVAKVLYRYIDDAALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTRDM
VRLEAEMANRAIWLSGRASHGVREEVLQTTFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLESAPGRIVAEVGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLVNVARTIAHDRIHWAVRQKQNLADLGARLAAISTALGLARGSNKHSIPNTI
KGAKPMVSGITTFPKSLDHAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAVFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGVLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1517 | GenBank | WP_128409206 |
| Name | traG_ELG73_RS33515_pSM16_Rh07 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69653.97 Da Isoelectric Point: 9.3813
>WP_128409206.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSSFMKTVPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDVGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGRAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSSFMKTVPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDVGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGRAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 175659..197692
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELG73_RS33375 (ELG73_33375) | 171189..172211 | - | 1023 | WP_128408794 | plasmid partitioning protein RepB | - |
| ELG73_RS33380 (ELG73_33380) | 172208..173422 | - | 1215 | WP_130784921 | plasmid partitioning protein RepA | - |
| ELG73_RS39215 | 174334..174546 | - | 213 | WP_245458276 | hypothetical protein | - |
| ELG73_RS33395 (ELG73_33395) | 175024..175662 | + | 639 | WP_063473646 | acyl-homoserine-lactone synthase | - |
| ELG73_RS33400 (ELG73_33400) | 175659..176636 | + | 978 | WP_130784875 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELG73_RS33405 (ELG73_33405) | 176626..177009 | + | 384 | WP_128408797 | TrbC/VirB2 family protein | virB2 |
| ELG73_RS33410 (ELG73_33410) | 177002..177301 | + | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELG73_RS33415 (ELG73_33415) | 177312..179768 | + | 2457 | Protein_176 | conjugal transfer protein TrbE | - |
| ELG73_RS33420 (ELG73_33420) | 179740..180543 | + | 804 | WP_128408799 | P-type conjugative transfer protein TrbJ | virB5 |
| ELG73_RS33425 (ELG73_33425) | 180540..180740 | + | 201 | WP_128408800 | entry exclusion protein TrbK | - |
| ELG73_RS33430 (ELG73_33430) | 180734..181915 | + | 1182 | WP_130784876 | P-type conjugative transfer protein TrbL | virB6 |
| ELG73_RS33435 (ELG73_33435) | 181936..182598 | + | 663 | WP_128408802 | conjugal transfer protein TrbF | virB8 |
| ELG73_RS33440 (ELG73_33440) | 182616..183428 | + | 813 | WP_130784877 | P-type conjugative transfer protein TrbG | virB9 |
| ELG73_RS33445 (ELG73_33445) | 183432..183872 | + | 441 | WP_128408804 | conjugal transfer protein TrbH | - |
| ELG73_RS33450 (ELG73_33450) | 183884..185182 | + | 1299 | WP_130784878 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELG73_RS33455 (ELG73_33455) | 185491..186195 | + | 705 | WP_128408806 | autoinducer binding domain-containing protein | - |
| ELG73_RS33460 (ELG73_33460) | 186180..186521 | - | 342 | WP_128408807 | transcriptional repressor TraM | - |
| ELG73_RS33465 (ELG73_33465) | 186747..186989 | + | 243 | WP_128408808 | helix-turn-helix transcriptional regulator | - |
| ELG73_RS33470 (ELG73_33470) | 187103..187735 | + | 633 | WP_128408809 | DUF433 domain-containing protein | - |
| ELG73_RS33475 (ELG73_33475) | 187732..188106 | + | 375 | WP_128408810 | DUF5615 family PIN-like protein | - |
| ELG73_RS33480 (ELG73_33480) | 188463..189178 | + | 716 | Protein_189 | Crp/Fnr family transcriptional regulator | - |
| ELG73_RS33485 (ELG73_33485) | 189323..189943 | - | 621 | WP_128408811 | TraH family protein | virB1 |
| ELG73_RS33490 (ELG73_33490) | 189961..191127 | - | 1167 | WP_128408812 | conjugal transfer protein TraB | - |
| ELG73_RS33495 (ELG73_33495) | 191117..191629 | - | 513 | WP_245458277 | conjugative transfer signal peptidase TraF | - |
| ELG73_RS33500 (ELG73_33500) | 191677..195003 | - | 3327 | WP_130784879 | Ti-type conjugative transfer relaxase TraA | - |
| ELG73_RS33505 (ELG73_33505) | 195258..195551 | + | 294 | WP_128409207 | conjugal transfer protein TraC | - |
| ELG73_RS33510 (ELG73_33510) | 195556..195771 | + | 216 | WP_011654561 | type IV conjugative transfer system coupling protein TraD | - |
| ELG73_RS33515 (ELG73_33515) | 195758..197692 | + | 1935 | WP_128409206 | Ti-type conjugative transfer system protein TraG | virb4 |
| ELG73_RS33525 (ELG73_33525) | 198132..198434 | + | 303 | Protein_197 | thermonuclease family protein | - |
| ELG73_RS33530 (ELG73_33530) | 198504..199583 | + | 1080 | WP_130784880 | IS630 family transposase | - |
| ELG73_RS33535 (ELG73_33535) | 199580..199729 | + | 150 | Protein_199 | thermonuclease family protein | - |
| ELG73_RS33540 (ELG73_33540) | 199961..200275 | - | 315 | WP_128408836 | hypothetical protein | - |
| ELG73_RS39575 (ELG73_33545) | 200719..201027 | + | 309 | WP_130784881 | WGR domain-containing protein | - |
| ELG73_RS39225 | 201196..201339 | + | 144 | WP_245488070 | hypothetical protein | - |
| ELG73_RS38545 | 201561..201722 | - | 162 | WP_018516739 | hypothetical protein | - |
| ELG73_RS33555 (ELG73_33555) | 202023..202334 | - | 312 | WP_127431460 | DUF736 domain-containing protein | - |
Host bacterium
| ID | 2997 | GenBank | NZ_SIKC01000007 |
| Plasmid name | pSM16_Rh07 | Incompatibility group | - |
| Plasmid size | 273515 bp | Coordinate of oriT [Strand] | 195075..195115 [-] |
| Host baterium | Rhizobium leguminosarum strain SM16 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifT, nodJ, nodI, nodC, nodA, nodD, nodF, nodE, nodM, fixG, fixQ, fixO, fixN |
| Anti-CRISPR | - |