Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102551 |
| Name | oriT_pSM52_Rh13 |
| Organism | Rhizobium leguminosarum strain SM52 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SILH01000005 (103706..103755 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pSM52_Rh13
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGCTGAAGGG
GGATCCAAGGGCGCAATTATACGTCGCTGACGCGACGCCCTGCTGAAGGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file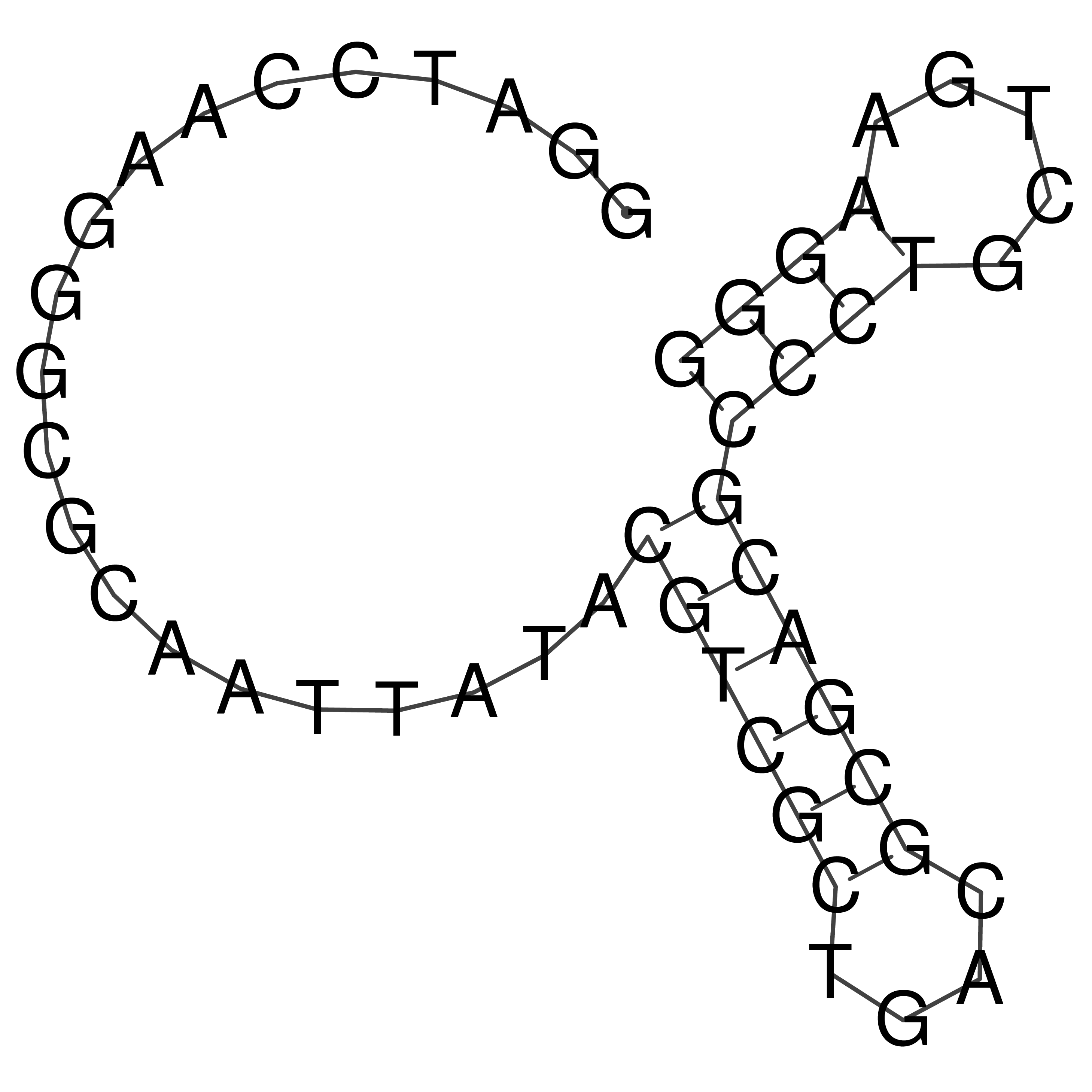
Relaxase
| ID | 1902 | GenBank | WP_011654745 |
| Name | traA_ELH04_RS35635_pSM52_Rh13 |
UniProt ID | Q1M997 |
| Length | 1197 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1197 a.a. Molecular weight: 131887.62 Da Isoelectric Point: 9.1379
>WP_011654745.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPETAPDWLRSMIADR
SVSGASEAFWNKVEAFEKRADAQLAKDVTIALPVELSNDQNIALVRDFVERHITAKGMVADWVYHDALGN
PHIHLMTTLRPLTEDGFGAKKVAVLAPAGKPVRNDAGKIVYELWAGSTDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKGDNKTGWGEEKVALERLELQEERRAENARRIQRNPEI
VLDLITREKSVFDERDIAKILYRYIDDAALFQNLMARILQSPQTLRLDRERMDLVTGVRAPSKYTTRELI
RLEAQMANQAIWLSQRSSHGVRHAVLSGVFSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKA
ARQAWEAAGYRVVGGALAGKAAEGLEKEAGIASRTLSAWELRWDQERDRLDEKSVFVLDEAGMVSSRQMA
RFVEAVTVSGAKLVLVGDPEQLQPIEAGAAFRAISGRIGYAELETIYRQREQWMRDASLDLARGNVSAAL
DAYAQRDMVRTGWTRDEAITALIADWDHEYDPAKSTLILAHRRIDVRLLNEMARSKLVERGLIEAGHAFK
TEDGTRQLAAGDQIVFLKNEGSLGVKNGMLARVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYAT
TVHKSQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRD
SYYVTLGFADGQERTVWGVDLARAMDASGARIGDRIGLKHVGSQRVTLPDGTEVDRNSWKVVPVEELAMA
RLHERLSRPGSKETTLDYQDASHYRAALRFAEARSLHLMNVARTIAHDQLQWTIRQSSKLAELGARLVAV
AAKLGLGGAKSTVSTTSAIKEAKPMVSGTTTFPRSIGQAAEDKLSADPGLKASWQEVSARFHHVFADPQA
AFKTVNVDAMLANGTVAATTIVQIAEQPESFGALKGKTGLFAGSAEKRAHDTALVNAPALARDLQGFIAK
RAEAASRYEDEERAVRTKLSLDIPALSASAKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAV
SERFGERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQ
TKGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYSRKQGLLHEEFVIPETAPDWLRSMIADR
SVSGASEAFWNKVEAFEKRADAQLAKDVTIALPVELSNDQNIALVRDFVERHITAKGMVADWVYHDALGN
PHIHLMTTLRPLTEDGFGAKKVAVLAPAGKPVRNDAGKIVYELWAGSTDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKGDNKTGWGEEKVALERLELQEERRAENARRIQRNPEI
VLDLITREKSVFDERDIAKILYRYIDDAALFQNLMARILQSPQTLRLDRERMDLVTGVRAPSKYTTRELI
RLEAQMANQAIWLSQRSSHGVRHAVLSGVFSRHDRLSDEQKTAIEHVAGPERIAAVIGRAGAGKTTMMKA
ARQAWEAAGYRVVGGALAGKAAEGLEKEAGIASRTLSAWELRWDQERDRLDEKSVFVLDEAGMVSSRQMA
RFVEAVTVSGAKLVLVGDPEQLQPIEAGAAFRAISGRIGYAELETIYRQREQWMRDASLDLARGNVSAAL
DAYAQRDMVRTGWTRDEAITALIADWDHEYDPAKSTLILAHRRIDVRLLNEMARSKLVERGLIEAGHAFK
TEDGTRQLAAGDQIVFLKNEGSLGVKNGMLARVVDAQPGRIVAEIGNGEDRRRVVVEQRFYANVDHGYAT
TVHKSQGATVDRVKVLASSTLDRHLSYVAMTRHRETAELYVGLEEFAQRRGGVLIAHGEAPYEHKPGNRD
SYYVTLGFADGQERTVWGVDLARAMDASGARIGDRIGLKHVGSQRVTLPDGTEVDRNSWKVVPVEELAMA
RLHERLSRPGSKETTLDYQDASHYRAALRFAEARSLHLMNVARTIAHDQLQWTIRQSSKLAELGARLVAV
AAKLGLGGAKSTVSTTSAIKEAKPMVSGTTTFPRSIGQAAEDKLSADPGLKASWQEVSARFHHVFADPQA
AFKTVNVDAMLANGTVAATTIVQIAEQPESFGALKGKTGLFAGSAEKRAHDTALVNAPALARDLQGFIAK
RAEAASRYEDEERAVRTKLSLDIPALSASAKQVLERVRDAIDRNDIPAGLEFALADKMVKAELEGFAKAV
SERFGERTFLPLAAKTADGKAFEVASAGMQPAQKNELRSAWDTIRTVQQLAAHERTAVALKQAEAIRQTQ
TKGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q1M997 |
T4CP
| ID | 1513 | GenBank | WP_011654760 |
| Name | t4cp2_ELH04_RS35550_pSM52_Rh13 |
UniProt ID | _ |
| Length | 811 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 811 a.a. Molecular weight: 90895.34 Da Isoelectric Point: 5.8208
>WP_011654760.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERARKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGASLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPGQWLIERGINRNE
MVALRTFRSTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSESATDFERNELSRQINSILSRLG
TGWMIQVEAVRVPTTEYPSAERSHFPDAVTLLIDHERRRHFGQERGHFESRHALILTYRPPERRRSGLTR
YIYSDNESRSAKYADTALDAFRRSIREIEQYLGNVLSVQRMQTREVSERDGARVARYDELFQFIRFAITG
ENHPVRLPEIPMYLDWLATAELEHGLTPRVEGRFLGVIAIDGLPAESWPGILNSLNLMPLTYRWSSRFIF
LDAEEAKQRLERARKKWQQKVRPFFDQLFQTQSRSVDQDAMAMVAETEDAIAEASSQLVAYGYYTPVIVL
FDESRSALQEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYCNIREPLINTRNLADLVPLNSVWS
GQPFAPCPFYPPDAPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFCRYEKAQVFA
FDKGNSMLTLTLGVGGDHYEIGGESGEGASLAFCPLSELSTDADRAWASEWIETLVSLQGVTIAPDHRNA
ISRQIGLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGLLLDAEEDGLTLGRFQCFEIEQLMNMGE
RNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVILATQSISDAERS
GIIDVLKESCPTKICLPNGAARETGTREFYERIGFNARQIEIVANAIPKREYYVTSPDGRRLFDMSLGPV
TLSFVGASGKADLARIRALSSTHGLEWPGQWLIERGINRNE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1514 | GenBank | WP_011654742 |
| Name | traG_ELH04_RS35650_pSM52_Rh13 |
UniProt ID | _ |
| Length | 650 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 650 a.a. Molecular weight: 70597.42 Da Isoelectric Point: 9.7031
>WP_011654742.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTPKRILVAGIPAVAMVAIALTFPGIERWLSAFGTTDQAKLMLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLGVIAVAALREGSRLLTFASQVPARRTLLSYADPSTIIGAGTALLVTFFALRVARMG
NAAFTRSEPRRIRGKRALHGEADWMTMQEAEKLFPETGGIVIGERYRVDRDDPAATSFRPGEPTSWGRGG
SPPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAQSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KHQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYANYG
ALVSGSSFSTEALADGETDVFINIDLKALETHAGLARVIIGSFLNAIYHRDGAIKGRALFLLDEVARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQVSRSFQSRGSSRTRSKQLASRQLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRSDMKSCVG
ENRFQQRGDRPEVSASAEPN
MTPKRILVAGIPAVAMVAIALTFPGIERWLSAFGTTDQAKLMLGRIGLALPYALAGACGVMFLFGTKGSI
NVKTSGWSVAAGGLGVIAVAALREGSRLLTFASQVPARRTLLSYADPSTIIGAGTALLVTFFALRVARMG
NAAFTRSEPRRIRGKRALHGEADWMTMQEAEKLFPETGGIVIGERYRVDRDDPAATSFRPGEPTSWGRGG
SPPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLVVLDPSNEVAPMVMEHRRKAGRRVIVLD
PKNAQSGFNALDWIGRHGGTKEEDIAAVASWIMSDSGRATGVRDDFFRASGLQLLTAMIADVCLSGHTDE
KHQTLRQVRANLSEPEPKLRARLQEIYDNSASDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYANYG
ALVSGSSFSTEALADGETDVFINIDLKALETHAGLARVIIGSFLNAIYHRDGAIKGRALFLLDEVARLGY
MRVLETARDAGRKYGITLTMIYQSIGQLRETYGGRDASSKWFESASWISFAAINDPETADYISRRCGTTT
VEIDQVSRSFQSRGSSRTRSKQLASRQLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRSDMKSCVG
ENRFQQRGDRPEVSASAEPN
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 83081..98309
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH04_RS35520 (ELH04_35520) | 79041..80258 | - | 1218 | WP_011654635 | plasmid replication protein RepC | - |
| ELH04_RS35525 (ELH04_35525) | 80433..81392 | - | 960 | WP_011654634 | plasmid partitioning protein RepB | - |
| ELH04_RS35530 (ELH04_35530) | 81516..82712 | - | 1197 | WP_011654633 | plasmid partitioning protein RepA | - |
| ELH04_RS35535 (ELH04_35535) | 83081..84046 | + | 966 | WP_011654763 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELH04_RS35540 (ELH04_35540) | 84036..84428 | + | 393 | WP_011654762 | TrbC/VirB2 family protein | virB2 |
| ELH04_RS35545 (ELH04_35545) | 84421..84720 | + | 300 | WP_011654761 | conjugal transfer protein TrbD | virB3 |
| ELH04_RS35550 (ELH04_35550) | 84731..87166 | + | 2436 | WP_011654760 | conjugal transfer protein TrbE | virb4 |
| ELH04_RS35555 (ELH04_35555) | 87159..87968 | + | 810 | WP_011654759 | P-type conjugative transfer protein TrbJ | virB5 |
| ELH04_RS35560 (ELH04_35560) | 87965..88165 | + | 201 | WP_028744197 | entry exclusion protein TrbK | - |
| ELH04_RS35565 (ELH04_35565) | 88159..89340 | + | 1182 | WP_011654758 | P-type conjugative transfer protein TrbL | virB6 |
| ELH04_RS35570 (ELH04_35570) | 89362..90024 | + | 663 | WP_011654757 | conjugal transfer protein TrbF | virB8 |
| ELH04_RS35575 (ELH04_35575) | 90040..90870 | + | 831 | WP_011654756 | P-type conjugative transfer protein TrbG | virB9 |
| ELH04_RS35580 (ELH04_35580) | 90874..91320 | + | 447 | WP_011654755 | conjugal transfer protein TrbH | - |
| ELH04_RS35585 (ELH04_35585) | 91336..92634 | + | 1299 | WP_011654754 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELH04_RS35590 (ELH04_35590) | 92836..93135 | + | 300 | WP_011654753 | transcriptional repressor TraM | - |
| ELH04_RS35595 (ELH04_35595) | 93136..93840 | - | 705 | WP_011654752 | autoinducer binding domain-containing protein | - |
| ELH04_RS35600 (ELH04_35600) | 94128..95747 | + | 1620 | WP_130749038 | hypothetical protein | - |
| ELH04_RS35605 (ELH04_35605) | 95737..96732 | + | 996 | WP_011654750 | alpha/beta hydrolase | - |
| ELH04_RS35610 (ELH04_35610) | 96875..97054 | + | 180 | WP_018010295 | hypothetical protein | - |
| ELH04_RS35615 (ELH04_35615) | 97051..97668 | + | 618 | WP_011654749 | hypothetical protein | - |
| ELH04_RS35620 (ELH04_35620) | 97692..98309 | - | 618 | WP_011654748 | TraH family protein | virB1 |
| ELH04_RS35625 (ELH04_35625) | 98325..99491 | - | 1167 | WP_011654747 | conjugal transfer protein TraB | - |
| ELH04_RS35630 (ELH04_35630) | 99481..100047 | - | 567 | WP_011654746 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 2995 | GenBank | NZ_SILH01000005 |
| Plasmid name | pSM52_Rh13 | Incompatibility group | - |
| Plasmid size | 147600 bp | Coordinate of oriT [Strand] | 103706..103755 [-] |
| Host baterium | Rhizobium leguminosarum strain SM52 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |