Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102549 |
| Name | oriT_pSM72_Rh01 |
| Organism | Rhizobium leguminosarum strain SM72 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SILY01000002 (708512..708540 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSM72_Rh01
AGGGCGCAATATACGTCGCTATCGCGACG
AGGGCGCAATATACGTCGCTATCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file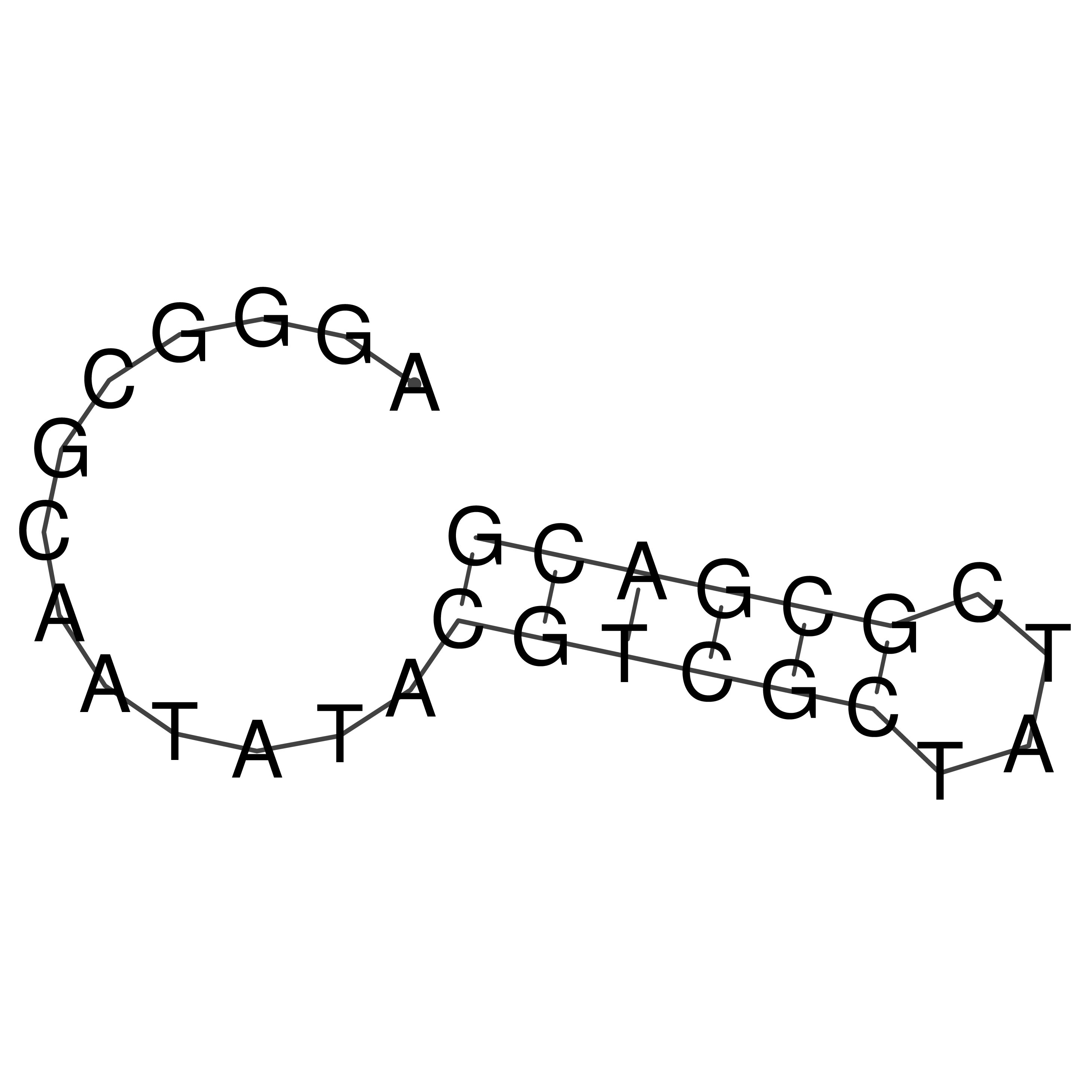
Relaxase
| ID | 1900 | GenBank | WP_130809845 |
| Name | traA_ELH21_RS28045_pSM72_Rh01 |
UniProt ID | _ |
| Length | 1543 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1543 a.a. Molecular weight: 170712.78 Da Isoelectric Point: 7.2717
>WP_130809845.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQMPTWLRTAIDG
RSVAGASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNFTSKGMVADWVYHDRDG
NPHIHLMTTLRPLTEEGFGQKKVAVTGDDGAPLRVVTPDRPKGRIVYRLWAGDKETMKAWKIAWADTANR
HLALAGHEVRLDGRSYAEQGLDGIAQTHLGPEKAALVRKGRELHFAPADLARRQEMADRLLADPTLLLKQ
LSNERSTFDERDIAKALHRTVDDPAVFANIRARLMASDDLIMLKPQQLDAETGKASEPAVFTTREVLRIE
YDMARSALLLAECRGFGVSDRVVAAAIRHVETRDTEKSFRLDPEQVDAVRHVTGDNGITAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELAWANGRELPERGNVLVIDEAGMVS
SQQLARVLDIAEQAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRHQWAREASRLFARGE
VEQGLDAYARHGHLIEAGTRDEVIGRIVGDWTDARRQAIRNSASAGNDGRLRGDELLVLAHTNEDVKRLN
EALRSVMSQEGALSENRAFRTERGAREFAVGDRIIFLENARFLEPRAPRLGPQHVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTLVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFEAKPEWGRKPRVDHAAGVTGALVESGQAKFRPNDEDADDSPYADVKTDDGTIHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVERVTIKVAIVDEQTGQKRFEEREVDRNVWTAKRIETSEARQQRIERESHRP
DLFKQLVERLSRSGAKTTTLDFESEAGYRAQAQDFARRRGLDHLSLAAAAVEESLSRRWAGIAERREQLA
KLWERAGVAFGFTIERERRVAYNEERAEPQPVLEADVVGSRYLIPPATGFARSAEEDALLAQLASPAWKE
REALLRPLLETIYRDPDAALVALNALSSDTSIEPRRLADDLAKAPERLGRLRGSDLIVDGRAARDERNAA
ITALTDLLPLARAHATEFRRNAERFESREQQRRAHMSLSIPTLSRKAMARLMEIEAVRRQGGDDAYKTAF
ALATEDRSIVQEVKAVSEALTARFGWSAFTAKADAVAERNMIERMPEDLTSISGEKLTRLFEAVKRFADE
QHLVERRDRSKIVAAASVDQGMETDKENAAVLPMLAAVTEFKTAVDEEARSRALATPLYRHQRAALAAVA
STIWRDPAGAVGKIEELLQKGFAGERIAAAVTNDPSAYGALRGSDRLMDRMLAAGRERKEAVQAMPEAAA
RLRSLGSAYVNVLDTERQAIAEERRRMAVAIPGLSTAAEDVLMRLTAEAKINGRKLSASAASLDPSIRRE
FAAVSSALDERFGRSAIIRGEKDLSNRVPPAQHRAFEVMQEKLKVLQQTVRRESSEQIISERRQRTTNRS
IDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGASELVHEELALPDQMPTWLRTAIDG
RSVAGASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVRDNFTSKGMVADWVYHDRDG
NPHIHLMTTLRPLTEEGFGQKKVAVTGDDGAPLRVVTPDRPKGRIVYRLWAGDKETMKAWKIAWADTANR
HLALAGHEVRLDGRSYAEQGLDGIAQTHLGPEKAALVRKGRELHFAPADLARRQEMADRLLADPTLLLKQ
LSNERSTFDERDIAKALHRTVDDPAVFANIRARLMASDDLIMLKPQQLDAETGKASEPAVFTTREVLRIE
YDMARSALLLAECRGFGVSDRVVAAAIRHVETRDTEKSFRLDPEQVDAVRHVTGDNGITAVVGLAGAGKS
TLLAAARVAWESDGRRVIGAALAGKAAEGLEDSSGIRSRTLAAWELAWANGRELPERGNVLVIDEAGMVS
SQQLARVLDIAEQAEVKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRHQWAREASRLFARGE
VEQGLDAYARHGHLIEAGTRDEVIGRIVGDWTDARRQAIRNSASAGNDGRLRGDELLVLAHTNEDVKRLN
EALRSVMSQEGALSENRAFRTERGAREFAVGDRIIFLENARFLEPRAPRLGPQHVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGATVDRTLVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFEAKPEWGRKPRVDHAAGVTGALVESGQAKFRPNDEDADDSPYADVKTDDGTIHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVERVTIKVAIVDEQTGQKRFEEREVDRNVWTAKRIETSEARQQRIERESHRP
DLFKQLVERLSRSGAKTTTLDFESEAGYRAQAQDFARRRGLDHLSLAAAAVEESLSRRWAGIAERREQLA
KLWERAGVAFGFTIERERRVAYNEERAEPQPVLEADVVGSRYLIPPATGFARSAEEDALLAQLASPAWKE
REALLRPLLETIYRDPDAALVALNALSSDTSIEPRRLADDLAKAPERLGRLRGSDLIVDGRAARDERNAA
ITALTDLLPLARAHATEFRRNAERFESREQQRRAHMSLSIPTLSRKAMARLMEIEAVRRQGGDDAYKTAF
ALATEDRSIVQEVKAVSEALTARFGWSAFTAKADAVAERNMIERMPEDLTSISGEKLTRLFEAVKRFADE
QHLVERRDRSKIVAAASVDQGMETDKENAAVLPMLAAVTEFKTAVDEEARSRALATPLYRHQRAALAAVA
STIWRDPAGAVGKIEELLQKGFAGERIAAAVTNDPSAYGALRGSDRLMDRMLAAGRERKEAVQAMPEAAA
RLRSLGSAYVNVLDTERQAIAEERRRMAVAIPGLSTAAEDVLMRLTAEAKINGRKLSASAASLDPSIRRE
FAAVSSALDERFGRSAIIRGEKDLSNRVPPAQHRAFEVMQEKLKVLQQTVRRESSEQIISERRQRTTNRS
IDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1510 | GenBank | WP_128410164 |
| Name | traG_ELH21_RS28030_pSM72_Rh01 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 70390.14 Da Isoelectric Point: 9.2899
>WP_128410164.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MALKGRLHPSLLLVFVPIAVTAITVYITGWRWTALAHGLSGKTEYWFLRAAPVPALLSGPLAGLLTVWAL
PLHRRRPIAIASFLCFLVVTGFYTLREYGRLVPSVERGVVTWNQALSYLDFIAVIGALAGFMAVAVAARI
SVVVPEAVKRARGGTFGDADWLSMAAAAKLFPRDGEIVVGERYRVDKETVHEAPFDPKDRSTWGQGGQAP
LLTYNQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVVEHRTRILGREVMVLDPA
NPIMGFNVLDGIEHSKRKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPECSGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSETAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVTGKTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFSRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKGGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVPGHSPIRCGRAIYFRRKDMDAQAK
ANRFVRR
MALKGRLHPSLLLVFVPIAVTAITVYITGWRWTALAHGLSGKTEYWFLRAAPVPALLSGPLAGLLTVWAL
PLHRRRPIAIASFLCFLVVTGFYTLREYGRLVPSVERGVVTWNQALSYLDFIAVIGALAGFMAVAVAARI
SVVVPEAVKRARGGTFGDADWLSMAAAAKLFPRDGEIVVGERYRVDKETVHEAPFDPKDRSTWGQGGQAP
LLTYNQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVVEHRTRILGREVMVLDPA
NPIMGFNVLDGIEHSKRKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPECSGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSETAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSSDIVTGKTDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFSRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVQG
RSRNIGWDTKGGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVPGHSPIRCGRAIYFRRKDMDAQAK
ANRFVRR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 340630..350189
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELH21_RS26035 (ELH21_26035) | 336709..337707 | + | 999 | WP_130809811 | ABC transporter substrate-binding protein | - |
| ELH21_RS26040 (ELH21_26040) | 337691..339466 | + | 1776 | WP_245512529 | adenylate/guanylate cyclase domain-containing protein | - |
| ELH21_RS26045 (ELH21_26045) | 339642..339995 | - | 354 | WP_128404288 | MarR family transcriptional regulator | - |
| ELH21_RS26050 (ELH21_26050) | 340087..340626 | + | 540 | WP_130809812 | hypothetical protein | - |
| ELH21_RS26055 (ELH21_26055) | 340630..341292 | + | 663 | WP_130809813 | transglycosylase SLT domain-containing protein | virB1 |
| ELH21_RS26060 (ELH21_26060) | 341289..341588 | + | 300 | WP_130660254 | TrbC/VirB2 family protein | virB2 |
| ELH21_RS26065 (ELH21_26065) | 341594..341932 | + | 339 | WP_130809814 | type IV secretion system protein VirB3 | virB3 |
| ELH21_RS26070 (ELH21_26070) | 341925..344291 | + | 2367 | WP_130809815 | VirB4 family type IV secretion system protein | virb4 |
| ELH21_RS26075 (ELH21_26075) | 344291..344989 | + | 699 | WP_130809816 | P-type DNA transfer protein VirB5 | virB5 |
| ELH21_RS26080 (ELH21_26080) | 344986..345219 | + | 234 | WP_130809817 | EexN family lipoprotein | - |
| ELH21_RS26085 (ELH21_26085) | 345223..346143 | + | 921 | WP_130666526 | type IV secretion system protein | virB6 |
| ELH21_RS26090 (ELH21_26090) | 346196..346480 | + | 285 | WP_130809818 | hypothetical protein | - |
| ELH21_RS26095 (ELH21_26095) | 346482..347153 | + | 672 | WP_130809819 | virB8 family protein | virB8 |
| ELH21_RS26100 (ELH21_26100) | 347150..348004 | + | 855 | WP_130809820 | P-type conjugative transfer protein VirB9 | virB9 |
| ELH21_RS26105 (ELH21_26105) | 348014..349186 | + | 1173 | WP_130809821 | type IV secretion system protein VirB10 | virB10 |
| ELH21_RS26110 (ELH21_26110) | 349194..350189 | + | 996 | WP_130809822 | P-type DNA transfer ATPase VirB11 | virB11 |
| ELH21_RS26120 (ELH21_26120) | 350906..351121 | - | 216 | WP_130766736 | DUF3072 domain-containing protein | - |
| ELH21_RS26125 (ELH21_26125) | 351369..351701 | - | 333 | Protein_321 | LysR substrate-binding domain-containing protein | - |
| ELH21_RS37680 (ELH21_26130) | 351708..351794 | + | 87 | Protein_322 | SDR family oxidoreductase | - |
| ELH21_RS26135 (ELH21_26135) | 352014..352922 | - | 909 | WP_130766952 | nucleotidyltransferase and HEPN domain-containing protein | - |
| ELH21_RS26140 (ELH21_26140) | 353266..353682 | + | 417 | WP_130660238 | UPF0158 family protein | - |
Host bacterium
| ID | 2993 | GenBank | NZ_SILY01000002 |
| Plasmid name | pSM72_Rh01 | Incompatibility group | - |
| Plasmid size | 1226126 bp | Coordinate of oriT [Strand] | 708512..708540 [+] |
| Host baterium | Rhizobium leguminosarum strain SM72 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | bphC |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | AcrIIA9 |