Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102548 |
| Name | oriT_pSM40_Rh07 |
| Organism | Rhizobium leguminosarum strain SM40 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIKV01000005 (67684..67724 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pSM40_Rh07
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
TCCAAGGGCGCAATTATACGTCGCTGGCGCGACGTCCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file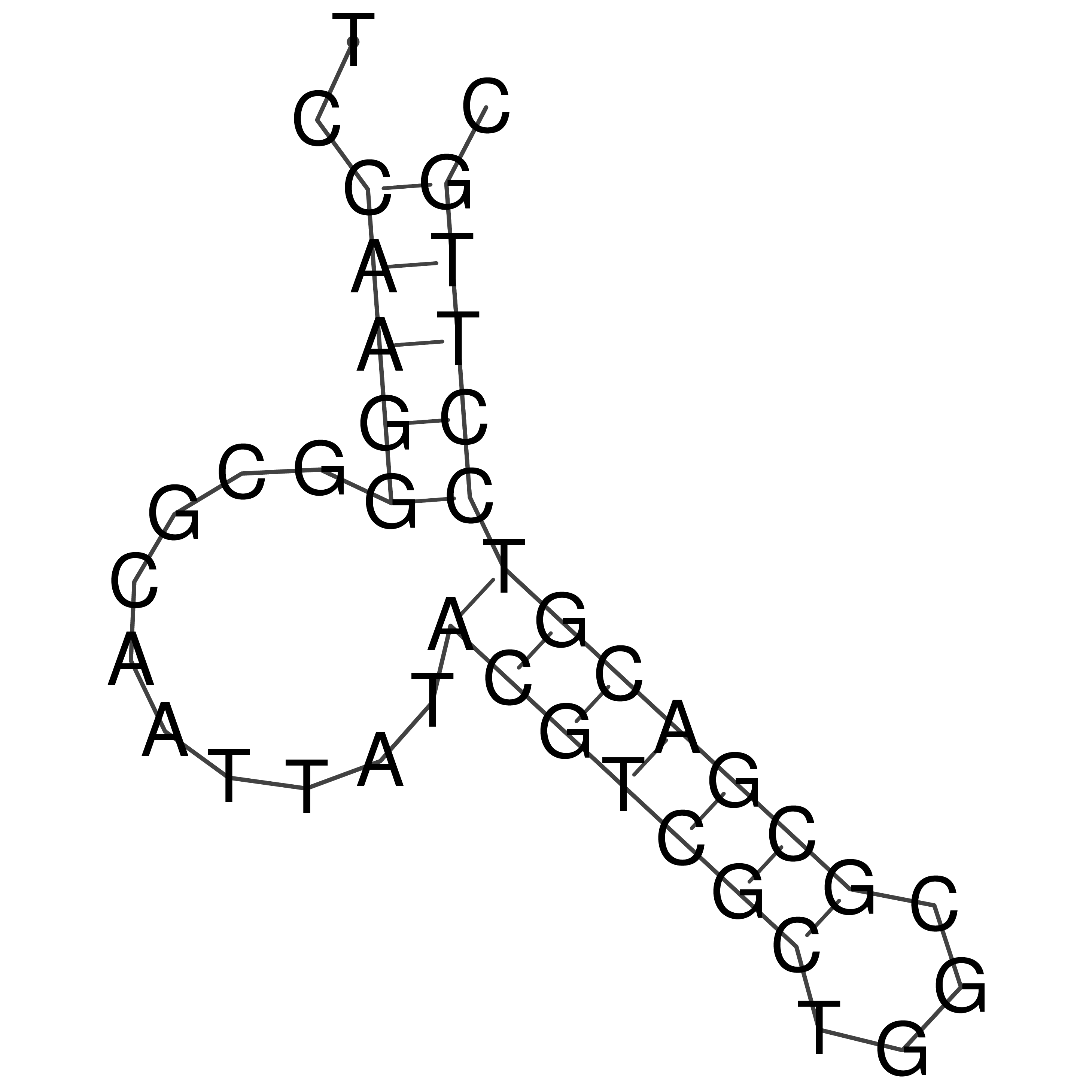
Relaxase
| ID | 1899 | GenBank | WP_130784879 |
| Name | traA_ELG92_RS35070_pSM40_Rh07 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122892.96 Da Isoelectric Point: 9.7033
>WP_130784879.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVCSMIADR
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKIALLGPDGKPVRNDAGKLVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKTQASAPKLERIELQGERRSENARRIERRPE
IVLDLITREKSVFDERDVAKVLYRYIDDAALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTRDM
VRLEAEMANRAIWLSGRASHGVREEVLQTTFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLESAPGRIVAEVGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLVNVARTIAHDRIHWAVRQKQNLADLGARLAAISTALGLARGSNKHSIPNTI
KGAKPMVSGITTFPKSLDHAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAVFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGVLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVCSMIADR
SVAGASEAFWNKVETFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKIALLGPDGKPVRNDAGKLVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
LEIHIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKTQASAPKLERIELQGERRSENARRIERRPE
IVLDLITREKSVFDERDVAKVLYRYIDDAALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTRDM
VRLEAEMANRAIWLSGRASHGVREEVLQTTFARHSRLSDEQRTAIEHVARATRIAAIIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQM
ALFVEAVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDLSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQVVFLKNEGSLGVKNGMLAKVLESAPGRIVAEVGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDKHLTYVAMTRHREDLAVYYGRRSFAKSGGLIPILSRRNAKETTLDYE
KASFYGQALRFAEARGLHLVNVARTIAHDRIHWAVRQKQNLADLGARLAAISTALGLARGSNKHSIPNTI
KGAKPMVSGITTFPKSLDHAVEDKLAADPGLKKQWEDVSTRFQLVYAQPEAVFKAINVDTMLKDQSVAQS
TLARIAGEPESFGALKGKTGVLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1508 | GenBank | WP_128409206 |
| Name | traG_ELG92_RS35055_pSM40_Rh07 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69653.97 Da Isoelectric Point: 9.3813
>WP_128409206.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSSFMKTVPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDVGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGRAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
MTINRIALVVMPGTLMILVVIGMTGVEQWLSGFGKTEAARLAWGRAGIALPYVASAAIGILLLFSSAGSI
NIKQAGWGVVAGCSGTILIAAIRETMRLSSFMKTVPADKTVLAFLDPATSIGASAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDVGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGRAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1509 | GenBank | WP_130794975 |
| Name | t4cp2_ELG92_RS35155_pSM40_Rh07 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91703.43 Da Isoelectric Point: 5.7027
>WP_130794975.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTLAIDAERRAHFAREQGHFESKHALVLTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLSVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMSMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPISPCPFYPPNAPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATSEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLETRGVHDAASLLKYV
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTLAIDAERRAHFAREQGHFESKHALVLTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLSVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMSMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAVRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPISPCPFYPPNAPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAMPKREYYVATSEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGRDWPIHWLETRGVHDAASLLKYV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 65107..87140
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELG92_RS35015 (ELG92_35030) | 60465..60776 | + | 312 | WP_127431460 | DUF736 domain-containing protein | - |
| ELG92_RS37745 | 61077..61238 | + | 162 | WP_018516739 | hypothetical protein | - |
| ELG92_RS38290 | 61460..61603 | - | 144 | WP_245488070 | hypothetical protein | - |
| ELG92_RS38610 (ELG92_35040) | 61772..62080 | - | 309 | WP_130784881 | WGR domain-containing protein | - |
| ELG92_RS35030 (ELG92_35045) | 62524..62838 | + | 315 | WP_128408836 | hypothetical protein | - |
| ELG92_RS35035 (ELG92_35050) | 63070..63219 | - | 150 | Protein_65 | thermonuclease family protein | - |
| ELG92_RS35040 (ELG92_35055) | 63216..64295 | - | 1080 | WP_130784880 | IS630 family transposase | - |
| ELG92_RS35045 (ELG92_35060) | 64365..64667 | - | 303 | Protein_67 | thermonuclease family protein | - |
| ELG92_RS35055 (ELG92_35070) | 65107..67041 | - | 1935 | WP_128409206 | Ti-type conjugative transfer system protein TraG | virb4 |
| ELG92_RS35060 (ELG92_35075) | 67028..67243 | - | 216 | WP_011654561 | type IV conjugative transfer system coupling protein TraD | - |
| ELG92_RS35065 (ELG92_35080) | 67248..67541 | - | 294 | WP_128409207 | conjugal transfer protein TraC | - |
| ELG92_RS35070 (ELG92_35085) | 67796..71122 | + | 3327 | WP_130784879 | Ti-type conjugative transfer relaxase TraA | - |
| ELG92_RS35075 (ELG92_35090) | 71170..71682 | + | 513 | WP_245458277 | conjugative transfer signal peptidase TraF | - |
| ELG92_RS35080 (ELG92_35095) | 71672..72838 | + | 1167 | WP_128408812 | conjugal transfer protein TraB | - |
| ELG92_RS35085 (ELG92_35100) | 72856..73476 | + | 621 | WP_128408811 | TraH family protein | virB1 |
| ELG92_RS35090 (ELG92_35105) | 73621..74336 | - | 716 | Protein_75 | Crp/Fnr family transcriptional regulator | - |
| ELG92_RS35095 (ELG92_35110) | 74693..75067 | - | 375 | WP_128408810 | DUF5615 family PIN-like protein | - |
| ELG92_RS35100 (ELG92_35115) | 75064..75696 | - | 633 | WP_128408809 | DUF433 domain-containing protein | - |
| ELG92_RS35105 (ELG92_35120) | 75810..76052 | - | 243 | WP_128408808 | helix-turn-helix transcriptional regulator | - |
| ELG92_RS35110 (ELG92_35125) | 76278..76619 | + | 342 | WP_128408807 | transcriptional repressor TraM | - |
| ELG92_RS35115 (ELG92_35130) | 76604..77308 | - | 705 | WP_128408806 | autoinducer binding domain-containing protein | - |
| ELG92_RS35120 (ELG92_35135) | 77617..78915 | - | 1299 | WP_130794973 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELG92_RS35125 (ELG92_35140) | 78927..79367 | - | 441 | WP_128408804 | conjugal transfer protein TrbH | - |
| ELG92_RS35130 (ELG92_35145) | 79371..80183 | - | 813 | WP_130784877 | P-type conjugative transfer protein TrbG | virB9 |
| ELG92_RS35135 (ELG92_35150) | 80201..80863 | - | 663 | WP_128408802 | conjugal transfer protein TrbF | virB8 |
| ELG92_RS35140 (ELG92_35155) | 80884..82065 | - | 1182 | WP_130784876 | P-type conjugative transfer protein TrbL | virB6 |
| ELG92_RS35145 (ELG92_35160) | 82059..82259 | - | 201 | WP_128408800 | entry exclusion protein TrbK | - |
| ELG92_RS35150 (ELG92_35165) | 82256..83059 | - | 804 | WP_128408799 | P-type conjugative transfer protein TrbJ | virB5 |
| ELG92_RS35155 (ELG92_35170) | 83031..85487 | - | 2457 | WP_130794975 | conjugal transfer protein TrbE | virb4 |
| ELG92_RS35160 (ELG92_35175) | 85498..85797 | - | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELG92_RS35165 (ELG92_35180) | 85790..86173 | - | 384 | WP_128408797 | TrbC/VirB2 family protein | virB2 |
| ELG92_RS35170 (ELG92_35185) | 86163..87140 | - | 978 | WP_130784875 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELG92_RS35175 (ELG92_35190) | 87137..87775 | - | 639 | WP_063473646 | acyl-homoserine-lactone synthase | - |
| ELG92_RS38300 | 88253..88465 | + | 213 | WP_245458276 | hypothetical protein | - |
| ELG92_RS35185 (ELG92_35200) | 88565..90676 | - | 2112 | WP_130790737 | recombinase family protein | - |
| ELG92_RS35190 (ELG92_35205) | 90969..91430 | - | 462 | WP_024323550 | helix-turn-helix domain-containing protein | - |
Host bacterium
| ID | 2992 | GenBank | NZ_SIKV01000005 |
| Plasmid name | pSM40_Rh07 | Incompatibility group | - |
| Plasmid size | 431811 bp | Coordinate of oriT [Strand] | 67684..67724 [+] |
| Host baterium | Rhizobium leguminosarum strain SM40 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, nodM, nodE, nodF, nodA, nodC, nodI, nodJ, nifT, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE |
| Anti-CRISPR | - |