Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102541 |
| Name | oriT_pSM34_Rh07 |
| Organism | Rhizobium leguminosarum strain SM34 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIKP01000006 (82650..82687 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pSM34_Rh07
GGATCCGAAGGGCGCAATTATACGTCGCTGCCGCGACG
GGATCCGAAGGGCGCAATTATACGTCGCTGCCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file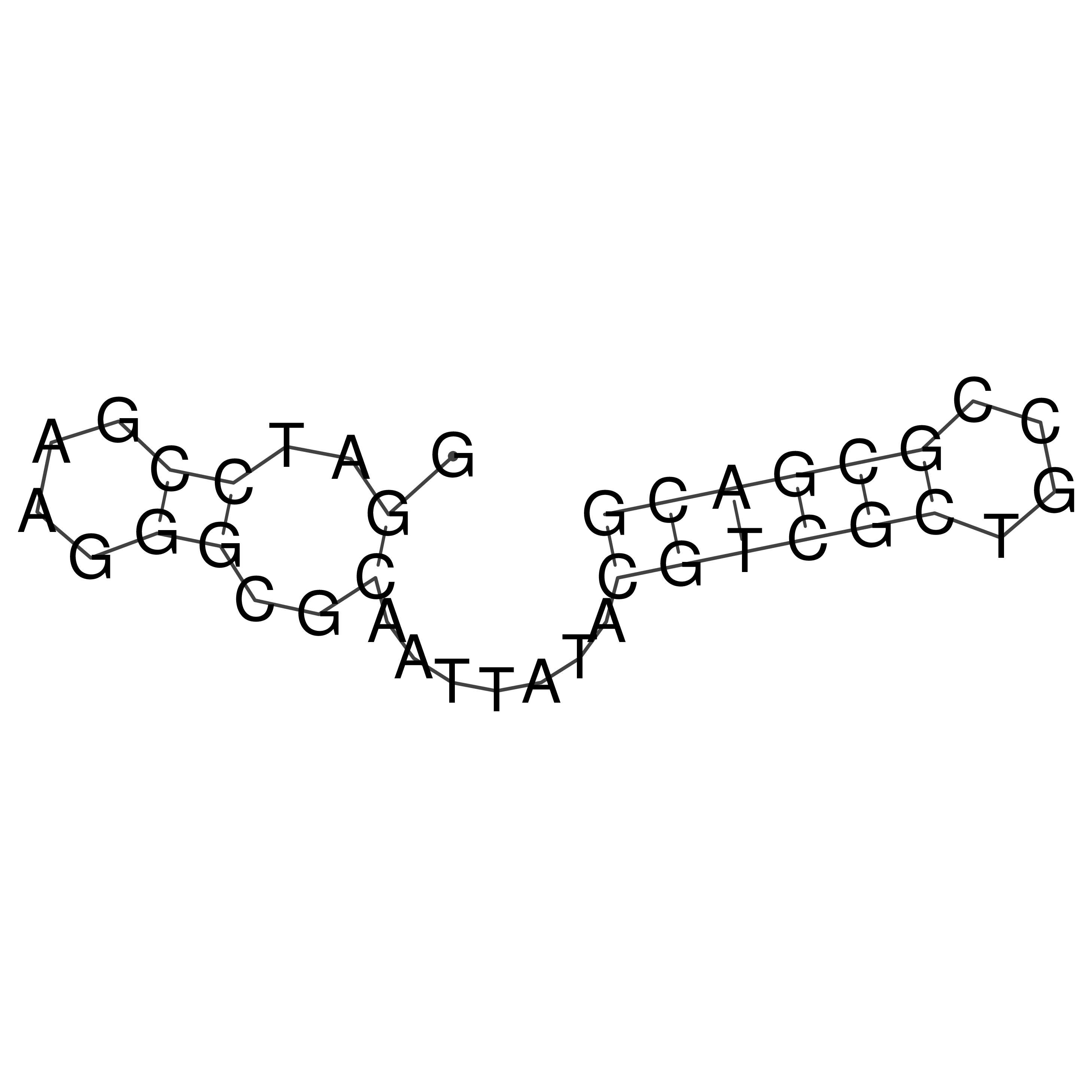
Relaxase
| ID | 1892 | GenBank | WP_130790745 |
| Name | traA_ELG86_RS37995_pSM34_Rh07 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123282.59 Da Isoelectric Point: 10.0651
>WP_130790745.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCANMEFEREARTIDYTRKQGLLHEEFVVPADAPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKSIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
RDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRRENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLAGRTSHGVREAMLRATFARHVRLSDEQRTAIEHIARATRIAAVIGRAGAGKTTMLK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIGSRTLSSWELRWNQGRNKFDHKTVFVLDEAGMVSSRQM
ALFVEAVSRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VEAYTVHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRKLNDMARAKLVERGVIADGSAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRVVAELGEGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KRALYRQALRFAEARGLHLMDVARTVAHDRLQWAVRQSSRLADLGARLAAIGAALGLVRGGNKQSIPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPKAAFKAINVDSMLKDQSVAQS
TLARIAGEPESFGALKGKTGLFASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCANMEFEREARTIDYTRKQGLLHEEFVVPADAPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKSIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
RDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRRENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLAGRTSHGVREAMLRATFARHVRLSDEQRTAIEHIARATRIAAVIGRAGAGKTTMLK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIGSRTLSSWELRWNQGRNKFDHKTVFVLDEAGMVSSRQM
ALFVEAVSRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VEAYTVHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRKLNDMARAKLVERGVIADGSAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRVVAELGEGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KRALYRQALRFAEARGLHLMDVARTVAHDRLQWAVRQSSRLADLGARLAAIGAALGLVRGGNKQSIPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPKAAFKAINVDSMLKDQSVAQS
TLARIAGEPESFGALKGKTGLFASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1497 | GenBank | WP_130790746 |
| Name | traG_ELG86_RS37980_pSM34_Rh07 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69472.74 Da Isoelectric Point: 9.2484
>WP_130790746.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYLASAAVGILFLFATAGAI
SIKQAGWGVVAGCSGTILIAALRETIRLSAFMKTVPADKTVLAFLDPATSIGANAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGGAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYLASAAVGILFLFATAGAI
SIKQAGWGVVAGCSGTILIAALRETIRLSAFMKTVPADKTVLAFLDPATSIGANAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGGAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1498 | GenBank | WP_130790739 |
| Name | t4cp2_ELG86_RS38080_pSM34_Rh07 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91814.32 Da Isoelectric Point: 5.5010
>WP_130790739.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPSNSPPLVQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRVLAFCPLSDLNSDVDRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEELMNMG
ERNLVPVLTYLFRRIERRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLETRGVHDAASLLRFE
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPSNSPPLVQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRVLAFCPLSDLNSDVDRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEELMNMG
ERNLVPVLTYLFRRIERRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLETRGVHDAASLLRFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 80074..102117
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELG86_RS39655 | 75946..76122 | - | 177 | WP_018482051 | hypothetical protein | - |
| ELG86_RS37955 (ELG86_37960) | 76573..76884 | + | 312 | WP_127431460 | DUF736 domain-containing protein | - |
| ELG86_RS39660 | 77185..77346 | + | 162 | WP_018516739 | hypothetical protein | - |
| ELG86_RS40230 | 77567..77710 | - | 144 | WP_245488070 | hypothetical protein | - |
| ELG86_RS40555 (ELG86_37970) | 77879..78187 | - | 309 | WP_130790747 | WGR domain-containing protein | - |
| ELG86_RS37970 (ELG86_37975) | 78631..78945 | + | 315 | WP_064251367 | hypothetical protein | - |
| ELG86_RS37975 (ELG86_37980) | 79188..79636 | - | 449 | Protein_80 | thermonuclease family protein | - |
| ELG86_RS37980 (ELG86_37985) | 80074..82008 | - | 1935 | WP_130790746 | Ti-type conjugative transfer system protein TraG | virb4 |
| ELG86_RS37985 (ELG86_37990) | 81995..82210 | - | 216 | WP_063473651 | type IV conjugative transfer system coupling protein TraD | - |
| ELG86_RS37990 (ELG86_37995) | 82215..82511 | - | 297 | WP_063473650 | conjugal transfer protein TraC | - |
| ELG86_RS37995 (ELG86_38000) | 82766..86092 | + | 3327 | WP_130790745 | Ti-type conjugative transfer relaxase TraA | - |
| ELG86_RS38000 (ELG86_38005) | 86089..86652 | + | 564 | WP_130790744 | conjugative transfer signal peptidase TraF | - |
| ELG86_RS38005 (ELG86_38010) | 86642..87808 | + | 1167 | WP_130817194 | conjugal transfer protein TraB | - |
| ELG86_RS38010 (ELG86_38015) | 87826..88446 | + | 621 | WP_130790742 | TraH family protein | virB1 |
| ELG86_RS38015 (ELG86_38020) | 88591..89313 | - | 723 | Protein_88 | Crp/Fnr family transcriptional regulator | - |
| ELG86_RS38020 (ELG86_38025) | 89670..90044 | - | 375 | WP_130790741 | DUF5615 family PIN-like protein | - |
| ELG86_RS38025 (ELG86_38030) | 90041..90673 | - | 633 | WP_128408809 | DUF433 domain-containing protein | - |
| ELG86_RS38030 (ELG86_38035) | 90787..91029 | - | 243 | WP_128408808 | helix-turn-helix transcriptional regulator | - |
| ELG86_RS38035 (ELG86_38040) | 91255..91596 | + | 342 | WP_128408807 | transcriptional repressor TraM | - |
| ELG86_RS38040 (ELG86_38045) | 91581..92285 | - | 705 | WP_128408806 | autoinducer binding domain-containing protein | - |
| ELG86_RS38045 (ELG86_38050) | 92594..93892 | - | 1299 | WP_130692286 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELG86_RS38050 (ELG86_38055) | 93904..94344 | - | 441 | WP_130692284 | conjugal transfer protein TrbH | - |
| ELG86_RS38055 (ELG86_38060) | 94348..95160 | - | 813 | WP_130759156 | P-type conjugative transfer protein TrbG | virB9 |
| ELG86_RS38060 (ELG86_38065) | 95178..95840 | - | 663 | WP_130692280 | conjugal transfer protein TrbF | virB8 |
| ELG86_RS38065 (ELG86_38070) | 95861..97042 | - | 1182 | WP_130759155 | P-type conjugative transfer protein TrbL | virB6 |
| ELG86_RS38070 (ELG86_38075) | 97036..97236 | - | 201 | WP_130759154 | entry exclusion protein TrbK | - |
| ELG86_RS38075 (ELG86_38080) | 97233..98036 | - | 804 | WP_130790740 | P-type conjugative transfer protein TrbJ | virB5 |
| ELG86_RS38080 (ELG86_38085) | 98008..100464 | - | 2457 | WP_130790739 | conjugal transfer protein TrbE | virb4 |
| ELG86_RS38085 (ELG86_38090) | 100475..100774 | - | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELG86_RS38090 (ELG86_38095) | 100767..101150 | - | 384 | WP_063473648 | TrbC/VirB2 family protein | virB2 |
| ELG86_RS38095 (ELG86_38100) | 101140..102117 | - | 978 | WP_130790738 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELG86_RS38100 (ELG86_38105) | 102114..102752 | - | 639 | WP_063473646 | acyl-homoserine-lactone synthase | - |
| ELG86_RS40240 | 103230..103442 | + | 213 | WP_245458276 | hypothetical protein | - |
| ELG86_RS38115 (ELG86_38120) | 104354..105568 | + | 1215 | WP_027691186 | plasmid partitioning protein RepA | - |
| ELG86_RS38120 (ELG86_38125) | 105565..106587 | + | 1023 | WP_128408794 | plasmid partitioning protein RepB | - |
Host bacterium
| ID | 2985 | GenBank | NZ_SIKP01000006 |
| Plasmid name | pSM34_Rh07 | Incompatibility group | - |
| Plasmid size | 327959 bp | Coordinate of oriT [Strand] | 82650..82687 [+] |
| Host baterium | Rhizobium leguminosarum strain SM34 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixN, fixO, fixQ, fixG, nodD, nodM, nodE, nodF, nodA, nodC, nodI, nodJ, nifT, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE |
| Anti-CRISPR | - |