Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102539 |
| Name | oriT_pSM37_Rh07 |
| Organism | Rhizobium leguminosarum strain SM37 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIKS01000005 (33073..33110 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pSM37_Rh07
GGATCCGAAGGGCGCAATTATACGTCGCTGCCGCGACG
GGATCCGAAGGGCGCAATTATACGTCGCTGCCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file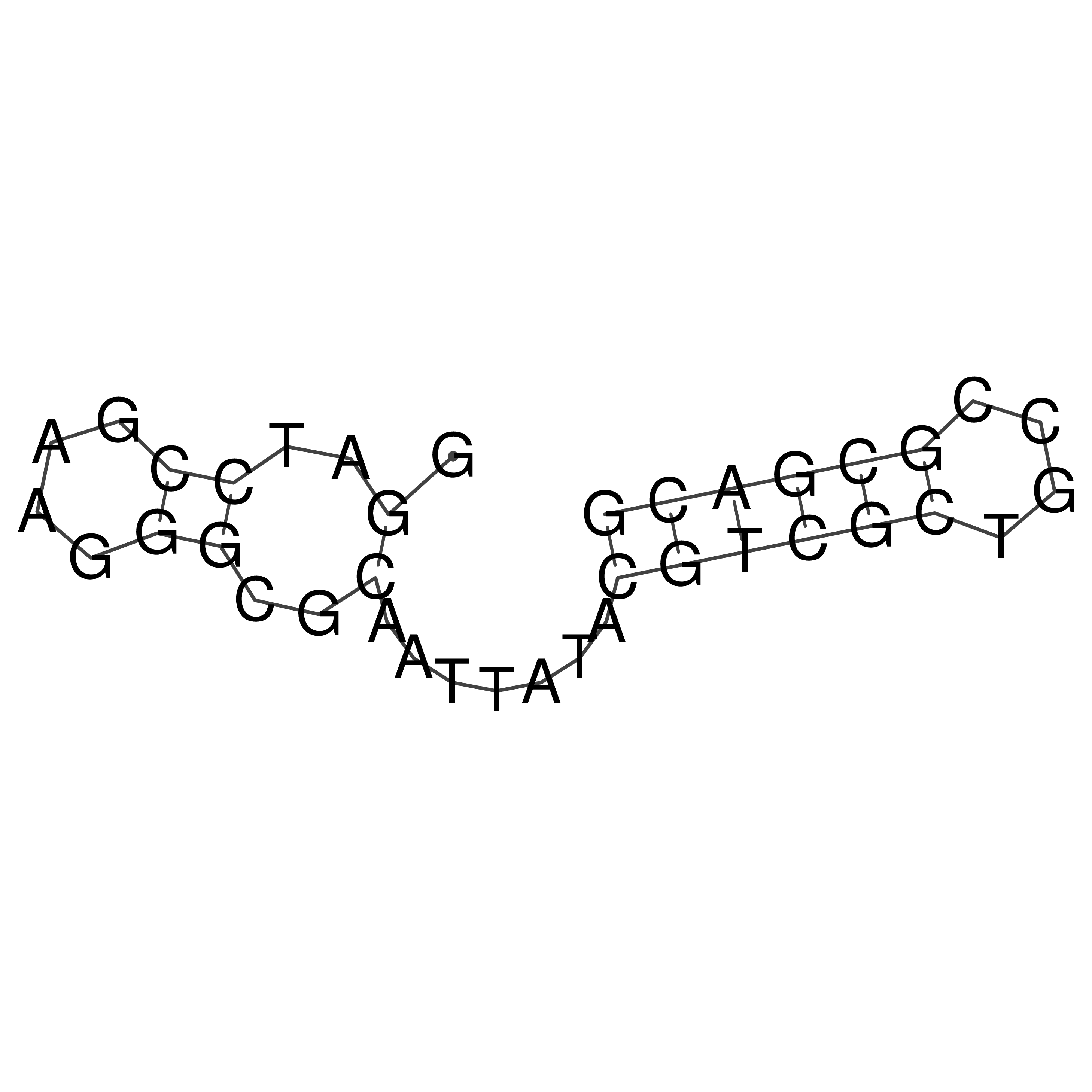
Relaxase
| ID | 1890 | GenBank | WP_130790745 |
| Name | traA_ELG89_RS31750_pSM37_Rh07 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123282.59 Da Isoelectric Point: 10.0651
>WP_130790745.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSVVARGSGRSAVLSAAYRHCANMEFEREARTIDYTRKQGLLHEEFVVPADAPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKSIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
RDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRRENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLAGRTSHGVREAMLRATFARHVRLSDEQRTAIEHIARATRIAAVIGRAGAGKTTMLK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIGSRTLSSWELRWNQGRNKFDHKTVFVLDEAGMVSSRQM
ALFVEAVSRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VEAYTVHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRKLNDMARAKLVERGVIADGSAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRVVAELGEGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KRALYRQALRFAEARGLHLMDVARTVAHDRLQWAVRQSSRLADLGARLAAIGAALGLVRGGNKQSIPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPKAAFKAINVDSMLKDQSVAQS
TLARIAGEPESFGALKGKTGLFASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCANMEFEREARTIDYTRKQGLLHEEFVVPADAPEWLRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVERHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKKVAVLGPDGKSIRNDAGKIVYQLWAGSTEDFNAFRDGWFACQNKHLALAG
RDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRRENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLAGRTSHGVREAMLRATFARHVRLSDEQRTAIEHIARATRIAAVIGRAGAGKTTMLK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIGSRTLSSWELRWNQGRNKFDHKTVFVLDEAGMVSSRQM
ALFVEAVSRAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VEAYTVHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRKLNDMARAKLVERGVIADGSAF
KTEDGTRMFAAGDQIVFLKNEGSLGVKNGMLAKVIEAAPGRVVAELGEGEHRRLVTIEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYE
KRALYRQALRFAEARGLHLMDVARTVAHDRLQWAVRQSSRLADLGARLAAIGAALGLVRGGNKQSIPNTI
KEAKPMVSGITTFPKSLDQAVEDKLAADPGLKKQWEDVSTRFQLVYAKPKAAFKAINVDSMLKDQSVAQS
TLARIAGEPESFGALKGKTGLFASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTADQKAEVQSAWNSMRTVQQLGSHERTTEALKQAETMRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1493 | GenBank | WP_130790739 |
| Name | t4cp2_ELG89_RS31665_pSM37_Rh07 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91814.32 Da Isoelectric Point: 5.5010
>WP_130790739.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPSNSPPLVQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRVLAFCPLSDLNSDVDRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEELMNMG
ERNLVPVLTYLFRRIERRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLETRGVHDAASLLRFE
MVALKRFRVTGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNEQSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSEDRCHFPDPVTRAIDAERRSHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANTLSIRRMETRETVERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKIENRFLGVVAIDGLPAESWPGILNSLDLIPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDSDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPSNSPPLVQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRVLAFCPLSDLNSDVDRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEELMNMG
ERNLVPVLTYLFRRIERRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATAIPKREYYVATPEGRRLFNMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLETRGVHDAASLLRFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1494 | GenBank | WP_130790746 |
| Name | traG_ELG89_RS31765_pSM37_Rh07 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69472.74 Da Isoelectric Point: 9.2484
>WP_130790746.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYLASAAVGILFLFATAGAI
SIKQAGWGVVAGCSGTILIAALRETIRLSAFMKTVPADKTVLAFLDPATSIGANAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGGAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYLASAAVGILFLFATAGAI
SIKQAGWGVVAGCSGTILIAALRETIRLSAFMKTVPADKTVLAFLDPATSIGANAALLCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLFPDAGGIVIGERYRVDKDSVGSQAFRADSAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSGHRGGAGRDVFLLD
PRKPDIGFNVLDWVGRFGGTKEEDIASIASWIMSDSGGVRGVRDDFFRASALQLLTALIADVCLSGHTDE
KDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGQIKGRALFLLDEVARLGY
MRIIETARDAGRKYGIALTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPRPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 13643..35686
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELG89_RS31630 (ELG89_31630) | 9353..9814 | + | 462 | WP_024323550 | helix-turn-helix domain-containing protein | - |
| ELG89_RS31635 (ELG89_31635) | 10107..12218 | + | 2112 | WP_130790737 | recombinase family protein | - |
| ELG89_RS37325 | 12318..12530 | - | 213 | WP_245458276 | hypothetical protein | - |
| ELG89_RS31645 (ELG89_31645) | 13008..13646 | + | 639 | WP_063473646 | acyl-homoserine-lactone synthase | - |
| ELG89_RS31650 (ELG89_31650) | 13643..14620 | + | 978 | WP_130790738 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELG89_RS31655 (ELG89_31655) | 14610..14993 | + | 384 | WP_063473648 | TrbC/VirB2 family protein | virB2 |
| ELG89_RS31660 (ELG89_31660) | 14986..15285 | + | 300 | WP_027681723 | conjugal transfer protein TrbD | virB3 |
| ELG89_RS31665 (ELG89_31665) | 15296..17752 | + | 2457 | WP_130790739 | conjugal transfer protein TrbE | virb4 |
| ELG89_RS31670 (ELG89_31670) | 17724..18527 | + | 804 | WP_130790740 | P-type conjugative transfer protein TrbJ | virB5 |
| ELG89_RS31675 (ELG89_31675) | 18524..18724 | + | 201 | WP_130759154 | entry exclusion protein TrbK | - |
| ELG89_RS31680 (ELG89_31680) | 18718..19899 | + | 1182 | WP_130759155 | P-type conjugative transfer protein TrbL | virB6 |
| ELG89_RS31685 (ELG89_31685) | 19920..20582 | + | 663 | WP_130692280 | conjugal transfer protein TrbF | virB8 |
| ELG89_RS31690 (ELG89_31690) | 20600..21412 | + | 813 | WP_130759156 | P-type conjugative transfer protein TrbG | virB9 |
| ELG89_RS31695 (ELG89_31695) | 21416..21856 | + | 441 | WP_130692284 | conjugal transfer protein TrbH | - |
| ELG89_RS31700 (ELG89_31700) | 21868..23166 | + | 1299 | WP_130692286 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELG89_RS31705 (ELG89_31705) | 23475..24179 | + | 705 | WP_128408806 | autoinducer binding domain-containing protein | - |
| ELG89_RS31710 (ELG89_31710) | 24164..24505 | - | 342 | WP_128408807 | transcriptional repressor TraM | - |
| ELG89_RS31715 (ELG89_31715) | 24731..24973 | + | 243 | WP_128408808 | helix-turn-helix transcriptional regulator | - |
| ELG89_RS31720 (ELG89_31720) | 25087..25719 | + | 633 | WP_128408809 | DUF433 domain-containing protein | - |
| ELG89_RS31725 (ELG89_31725) | 25716..26090 | + | 375 | WP_130790741 | DUF5615 family PIN-like protein | - |
| ELG89_RS31730 (ELG89_31730) | 26447..27169 | + | 723 | Protein_27 | Crp/Fnr family transcriptional regulator | - |
| ELG89_RS31735 (ELG89_31735) | 27314..27934 | - | 621 | WP_130790742 | TraH family protein | virB1 |
| ELG89_RS31740 (ELG89_31740) | 27952..29118 | - | 1167 | WP_130790743 | conjugal transfer protein TraB | - |
| ELG89_RS31745 (ELG89_31745) | 29108..29671 | - | 564 | WP_130790744 | conjugative transfer signal peptidase TraF | - |
| ELG89_RS31750 (ELG89_31750) | 29668..32994 | - | 3327 | WP_130790745 | Ti-type conjugative transfer relaxase TraA | - |
| ELG89_RS31755 (ELG89_31755) | 33249..33545 | + | 297 | WP_063473650 | conjugal transfer protein TraC | - |
| ELG89_RS31760 (ELG89_31760) | 33550..33765 | + | 216 | WP_063473651 | type IV conjugative transfer system coupling protein TraD | - |
| ELG89_RS31765 (ELG89_31765) | 33752..35686 | + | 1935 | WP_130790746 | Ti-type conjugative transfer system protein TraG | virb4 |
| ELG89_RS31770 (ELG89_31770) | 36124..36572 | + | 449 | Protein_35 | thermonuclease family protein | - |
| ELG89_RS31775 (ELG89_31775) | 36815..37129 | - | 315 | WP_064251367 | hypothetical protein | - |
| ELG89_RS37655 (ELG89_31780) | 37573..37881 | + | 309 | WP_130790747 | WGR domain-containing protein | - |
| ELG89_RS37335 | 38050..38193 | + | 144 | WP_245488070 | hypothetical protein | - |
| ELG89_RS36725 | 38414..38575 | - | 162 | WP_018516739 | hypothetical protein | - |
| ELG89_RS31790 (ELG89_31790) | 38876..39187 | - | 312 | WP_127431460 | DUF736 domain-containing protein | - |
| ELG89_RS36730 | 39638..39814 | + | 177 | WP_018482051 | hypothetical protein | - |
Host bacterium
| ID | 2983 | GenBank | NZ_SIKS01000005 |
| Plasmid name | pSM37_Rh07 | Incompatibility group | - |
| Plasmid size | 101260 bp | Coordinate of oriT [Strand] | 33073..33110 [-] |
| Host baterium | Rhizobium leguminosarum strain SM37 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD |
| Anti-CRISPR | - |