Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102532 |
| Name | oriT_pSM160_Rh08 |
| Organism | Rhizobium leguminosarum strain SM160 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIQL01000005 (93845..93882 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pSM160_Rh08
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file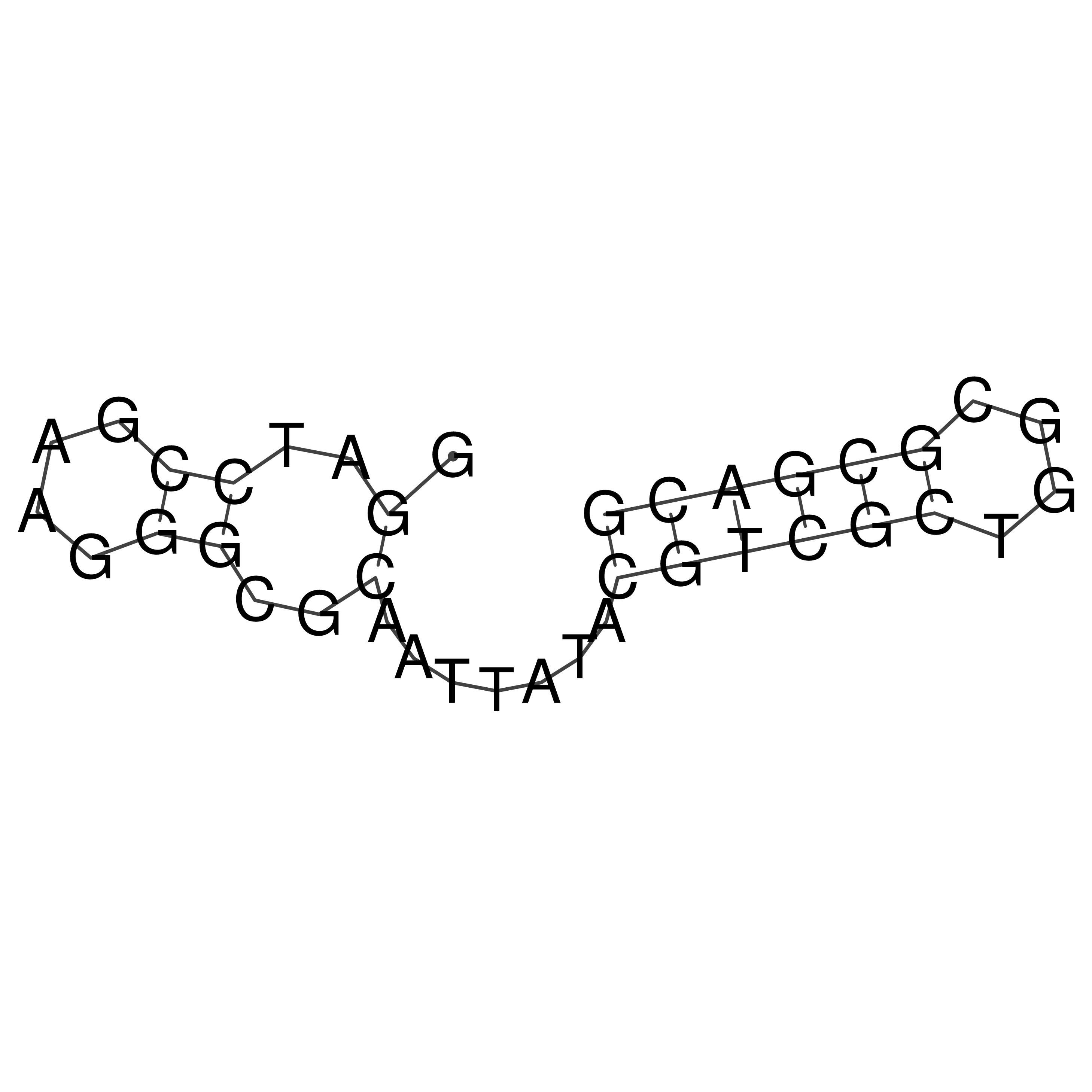
Relaxase
| ID | 1883 | GenBank | WP_130729904 |
| Name | traA_ELI38_RS35340_pSM160_Rh08 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122848.14 Da Isoelectric Point: 9.8768
>WP_130729904.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLIEDGFGSKKVAILGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSARASHGVREAVLHATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNRLDDKTVIVLDEAGMVSSRQM
ALLVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLARVLEAASGRIVAEIGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLAYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYQ
KSALYRQALRFAEARGLQLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIRDTI
KEAKPMVSGITTFPKSLDQAVEGKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKAINVDAMLTDQSVAQS
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLIEDGFGSKKVAILGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPAKYTTREM
IRLEAEMANRAIWLSARASHGVREAVLHATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNRLDDKTVIVLDEAGMVSSRQM
ALLVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLARVLEAASGRIVAEIGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLAYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYQ
KSALYRQALRFAEARGLQLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIRDTI
KEAKPMVSGITTFPKSLDQAVEGKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKAINVDAMLTDQSVAQS
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1482 | GenBank | WP_130729901 |
| Name | t4cp2_ELI38_RS35255_pSM160_Rh08 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91439.18 Da Isoelectric Point: 5.8944
>WP_130729901.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLESTRKKWQQKVRPFFEQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
HRNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAAPLLK
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLESTRKKWQQKVRPFFEQIFQTQSRSVDQDAMTMVVETQDAIAQASSRLVAYGYYTPVVVL
FDIDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQVGLMASASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
HRNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAAPLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1483 | GenBank | WP_130729905 |
| Name | traG_ELI38_RS35355_pSM160_Rh08 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69418.64 Da Isoelectric Point: 9.3575
>WP_130729905.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYGASAAIGILFLFASAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTKALNADKSVLAYLDPATAIGAGVALMCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLLPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGASG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEE
KEQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
MTINRIALVIVPAALMIPVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYGASAAIGILFLFASAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTKALNADKSVLAYLDPATAIGAGVALMCACFALRVALIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLLPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGASG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTEE
KEQTLRRVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFSTSDIAAGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 74574..88709
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI38_RS35220 (ELI38_35210) | 69901..71115 | - | 1215 | WP_130729900 | plasmid replication protein RepC | - |
| ELI38_RS35225 (ELI38_35215) | 71305..72342 | - | 1038 | WP_130657451 | plasmid partitioning protein RepB | - |
| ELI38_RS35230 (ELI38_35220) | 72339..73562 | - | 1224 | WP_065276670 | plasmid partitioning protein RepA | - |
| ELI38_RS35235 (ELI38_35225) | 73939..74577 | + | 639 | WP_130654277 | acyl-homoserine-lactone synthase | - |
| ELI38_RS35240 (ELI38_35230) | 74574..75551 | + | 978 | WP_130654278 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI38_RS35245 (ELI38_35235) | 75541..75924 | + | 384 | WP_065276633 | TrbC/VirB2 family protein | virB2 |
| ELI38_RS35250 (ELI38_35240) | 75917..76216 | + | 300 | WP_130654279 | conjugal transfer protein TrbD | virB3 |
| ELI38_RS35255 (ELI38_35245) | 76227..78677 | + | 2451 | WP_130729901 | conjugal transfer protein TrbE | virb4 |
| ELI38_RS35260 (ELI38_35250) | 78655..79452 | + | 798 | WP_130654281 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI38_RS35265 (ELI38_35255) | 79446..79622 | + | 177 | WP_130654282 | entry exclusion protein TrbK | - |
| ELI38_RS35270 (ELI38_35260) | 79627..80814 | + | 1188 | WP_130654283 | P-type conjugative transfer protein TrbL | virB6 |
| ELI38_RS35275 (ELI38_35265) | 80835..81497 | + | 663 | WP_130654284 | conjugal transfer protein TrbF | virB8 |
| ELI38_RS35280 (ELI38_35270) | 81515..82327 | + | 813 | WP_130729902 | P-type conjugative transfer protein TrbG | virB9 |
| ELI38_RS35285 (ELI38_35275) | 82331..82771 | + | 441 | WP_130654286 | conjugal transfer protein TrbH | - |
| ELI38_RS35290 (ELI38_35280) | 82783..84081 | + | 1299 | WP_130654287 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI38_RS35295 (ELI38_35285) | 84393..85097 | + | 705 | WP_130654288 | autoinducer binding domain-containing protein | - |
| ELI38_RS35300 (ELI38_35290) | 85101..85424 | - | 324 | WP_130654289 | transcriptional repressor TraM | - |
| ELI38_RS35305 (ELI38_35295) | 85646..85888 | + | 243 | WP_130654290 | helix-turn-helix transcriptional regulator | - |
| ELI38_RS37265 (ELI38_35300) | 85991..86422 | + | 432 | Protein_89 | hypothetical protein | - |
| ELI38_RS35315 (ELI38_35305) | 86513..87130 | + | 618 | WP_130654291 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| ELI38_RS35320 (ELI38_35310) | 87123..87989 | + | 867 | Protein_91 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| ELI38_RS35325 (ELI38_35315) | 88092..88709 | - | 618 | WP_130654292 | TraH family protein | virB1 |
| ELI38_RS35330 (ELI38_35320) | 88726..89889 | - | 1164 | WP_130729903 | conjugal transfer protein TraB | - |
| ELI38_RS35335 (ELI38_35325) | 89879..90442 | - | 564 | WP_130714114 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 2976 | GenBank | NZ_SIQL01000005 |
| Plasmid name | pSM160_Rh08 | Incompatibility group | - |
| Plasmid size | 107263 bp | Coordinate of oriT [Strand] | 93845..93882 [-] |
| Host baterium | Rhizobium leguminosarum strain SM160 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |