Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102531 |
| Name | oriT_pSM152A_Rh03 |
| Organism | Rhizobium leguminosarum strain SM152A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIPU01000004 (27756..27784 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSM152A_Rh03
AGGGCGCAATATACGTCGCTGTGGCGACG
AGGGCGCAATATACGTCGCTGTGGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file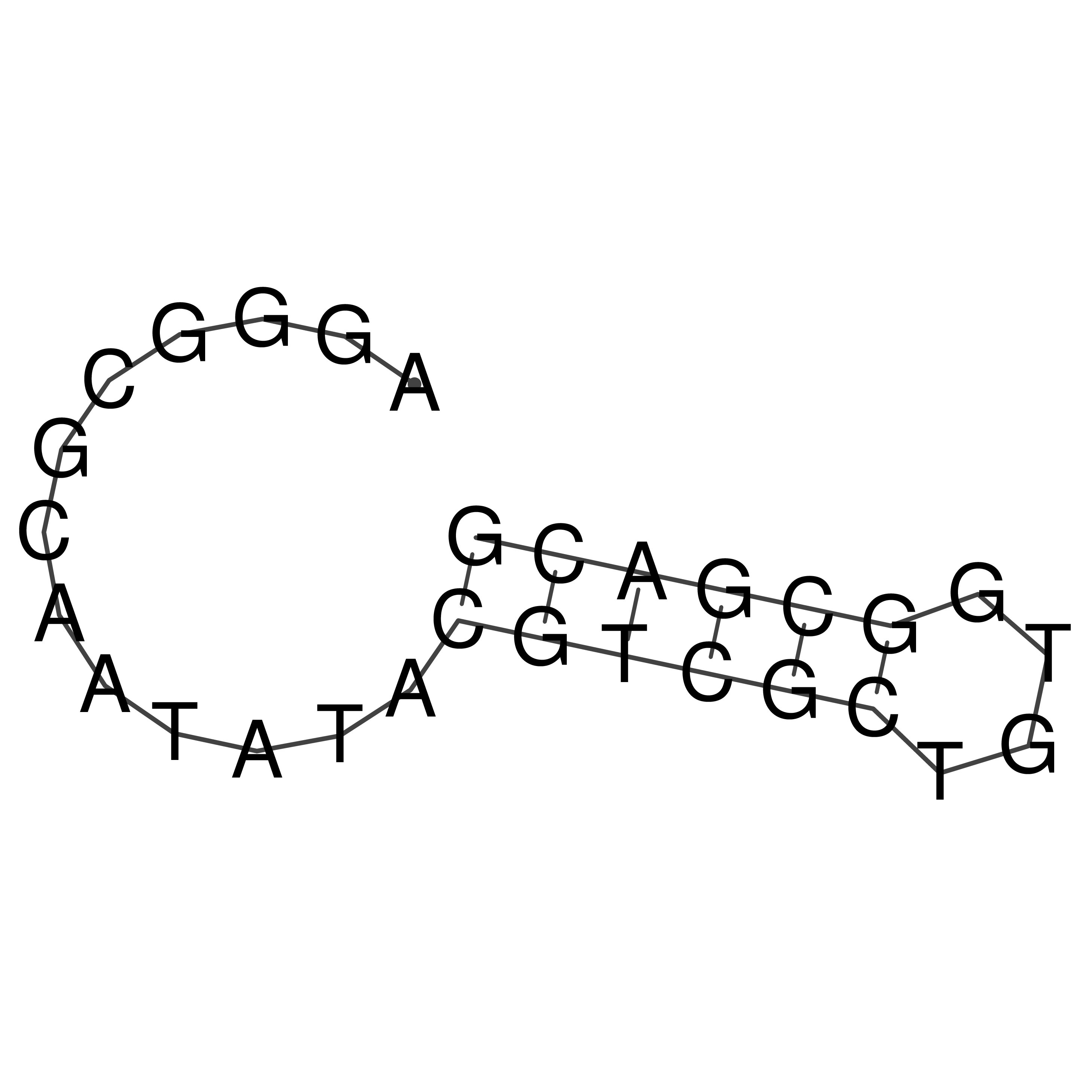
Relaxase
| ID | 1882 | GenBank | WP_130671197 |
| Name | traA_ELI21_RS29395_pSM152A_Rh03 |
UniProt ID | _ |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 170776.89 Da Isoelectric Point: 7.1939
>WP_130671197.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELSLPDQLPAWLRSAIEG
RSVAGASEALWNAVDAFEKRADAQLARELIIAVPEELTRGENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHVMTTLRPMTQEGFGPKKVAVTGEDGNSLRVVTPDRPNGKIVYKLWAGDKETMKVWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQRHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPELLLKQ
LGNERSTFDERDIAKALHRYVDDPAVFANIRARLMVSDDLVLLKPQQVDAETAKVAEPAVFTTRELLRIE
YDMARSAQVLSEHKGFAVSDRAVAAAIRHVETGDPEKSFRLDPEQVDAVRHVTRDNGIAAVVGLAGTGKS
TLLAAARVAWESGGSRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWANGHETLHRGDVVVVDEAGMVS
SQQMARVLDIVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQPWAREASRLFARGE
IEKGLDAYAQQGHLIEAGTRAETIDRIVADWSEARKRAIETAALKGNDGRLRGDELLVLAHTNQDVKRLN
EALRTVMTGEGALGENRSFQTERGGRDFAAGDRIIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISENSYRNVDHGYAATIHKSQGSTVERTLVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEARPEWGRKPRVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTEDGSVHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVERVKVQVPIVDEQTGQKRYEEREVDRNVWTAKRIETAAERLERIELESHRP
ELFKQLVERLSRSGAKTTTLDFESEAGYRQYADDFARRRGLDHLSQTAAGMEEGVSRRWAAIAEKREQLA
ELWERASVALGLAIERERRASYNEERIETQTVPDADAGRARYLIPSTTEFARSAEEDARLAQLGSPAWKD
REAILRPLLEKIYRDPDAALVALNALASDNTIEPRRLADDLAVAPDRLGRVRGSDLMVDGRAARDERSIA
IAALTELIPLVRAHATEFRRNAERFGVREQQRRAHMPLPIPALSKAAMARLVEIEAVRRQGGDDAYKTAF
ALATEDRSLVQEVKAVSEALTARFGWSAFTPNADALAERNMVERMPEDLASTRREKLTRLFEAVKHFAEE
QHLAERRDRSKIVAGASADQGMEAGKENAAVLPMLAAVTEFKTPIDEEARSRALATPLYRHQRAALAAVA
STIWRDPAGAIGKIEELLQKGFAGDRIAAAVTNDPSAYSALRGSDRLMDRMLAAGRERKDAVQALPEAAA
RLRALGSAYVNVLAAERHAIAEERRRMAVAIPGLSKAAEEALMRLTAEAKNNGRKRNASAVSLDPTIRQE
FAAVSRALDARFGRSAILRDEKDIINRIPPAQRRAFEAMQERFKVLQQTVRFESSEQIMSERRQRAVSRG
HGIDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELSLPDQLPAWLRSAIEG
RSVAGASEALWNAVDAFEKRADAQLARELIIAVPEELTRGENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHVMTTLRPMTQEGFGPKKVAVTGEDGNSLRVVTPDRPNGKIVYKLWAGDKETMKVWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQRHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPELLLKQ
LGNERSTFDERDIAKALHRYVDDPAVFANIRARLMVSDDLVLLKPQQVDAETAKVAEPAVFTTRELLRIE
YDMARSAQVLSEHKGFAVSDRAVAAAIRHVETGDPEKSFRLDPEQVDAVRHVTRDNGIAAVVGLAGTGKS
TLLAAARVAWESGGSRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWANGHETLHRGDVVVVDEAGMVS
SQQMARVLDIVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQPWAREASRLFARGE
IEKGLDAYAQQGHLIEAGTRAETIDRIVADWSEARKRAIETAALKGNDGRLRGDELLVLAHTNQDVKRLN
EALRTVMTGEGALGENRSFQTERGGRDFAAGDRIIFLENARFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISENSYRNVDHGYAATIHKSQGSTVERTLVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEARPEWGRKPRVDHAAGVTGELVETGQAKFRPNDEDADDSPYADVKTEDGSVHRLWGVSLPKA
LEEGGVSQGDTVTLRKDGVERVKVQVPIVDEQTGQKRYEEREVDRNVWTAKRIETAAERLERIELESHRP
ELFKQLVERLSRSGAKTTTLDFESEAGYRQYADDFARRRGLDHLSQTAAGMEEGVSRRWAAIAEKREQLA
ELWERASVALGLAIERERRASYNEERIETQTVPDADAGRARYLIPSTTEFARSAEEDARLAQLGSPAWKD
REAILRPLLEKIYRDPDAALVALNALASDNTIEPRRLADDLAVAPDRLGRVRGSDLMVDGRAARDERSIA
IAALTELIPLVRAHATEFRRNAERFGVREQQRRAHMPLPIPALSKAAMARLVEIEAVRRQGGDDAYKTAF
ALATEDRSLVQEVKAVSEALTARFGWSAFTPNADALAERNMVERMPEDLASTRREKLTRLFEAVKHFAEE
QHLAERRDRSKIVAGASADQGMEAGKENAAVLPMLAAVTEFKTPIDEEARSRALATPLYRHQRAALAAVA
STIWRDPAGAIGKIEELLQKGFAGDRIAAAVTNDPSAYSALRGSDRLMDRMLAAGRERKDAVQALPEAAA
RLRALGSAYVNVLAAERHAIAEERRRMAVAIPGLSKAAEEALMRLTAEAKNNGRKRNASAVSLDPTIRQE
FAAVSRALDARFGRSAILRDEKDIINRIPPAQRRAFEAMQERFKVLQQTVRFESSEQIMSERRQRAVSRG
HGIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1481 | GenBank | WP_130671194 |
| Name | traG_ELI21_RS29380_pSM152A_Rh03 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 70381.05 Da Isoelectric Point: 8.7930
>WP_130671194.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MALKGRLYPSLLLVFVPIALTAIAVYITGWRWPALAHGLSGKTEYWFLRVAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMASLFCFLLVAGFYTLREYGRLAPSVERGAVTWDHALSYLDFVAVIGALAGFIAVAAAARI
SVVVPEPVKRAKGGAFGDADWLSMSAAARLFPPDGEIVVGERYRVDKEVVHELPFDPNDRSTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPITGFNVLDGIEHSQRKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSDKKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFSRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNISAQCGEMTVEVQG
RSRNIGWDTKGGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDAQAK
ANRFVRT
MALKGRLYPSLLLVFVPIALTAIAVYITGWRWPALAHGLSGKTEYWFLRVAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMASLFCFLLVAGFYTLREYGRLAPSVERGAVTWDHALSYLDFVAVIGALAGFIAVAAAARI
SVVVPEPVKRAKGGAFGDADWLSMSAAARLFPPDGEIVVGERYRVDKEVVHELPFDPNDRSTWGQGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVLPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPITGFNVLDGIEHSQRKEEDIVGIAHMLLSESLRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSESAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSDKKDVFLNIPASILRSYPGIGRVIIGSLINAMVQADGGFSRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNISAQCGEMTVEVQG
RSRNIGWDTKGGGTRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDAQAK
ANRFVRT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 300664..310267
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI21_RS30680 (ELI21_30685) | 296519..297020 | - | 502 | Protein_277 | transposase | - |
| ELI21_RS30685 (ELI21_30690) | 297174..297539 | - | 366 | WP_130671356 | GtrA family protein | - |
| ELI21_RS30690 (ELI21_30695) | 297536..298240 | - | 705 | WP_130671357 | HAD family hydrolase | - |
| ELI21_RS30695 (ELI21_30700) | 298237..299212 | - | 976 | Protein_280 | UbiA prenyltransferase family protein | - |
| ELI21_RS30705 (ELI21_30710) | 299676..300029 | - | 354 | WP_130671358 | MarR family transcriptional regulator | - |
| ELI21_RS30710 (ELI21_30715) | 300121..300660 | + | 540 | WP_130671359 | hypothetical protein | - |
| ELI21_RS30715 (ELI21_30720) | 300664..301326 | + | 663 | WP_130671360 | lytic transglycosylase domain-containing protein | virB1 |
| ELI21_RS30720 (ELI21_30725) | 301323..301622 | + | 300 | WP_130671361 | TrbC/VirB2 family protein | virB2 |
| ELI21_RS30725 (ELI21_30730) | 301628..301966 | + | 339 | WP_130671362 | type IV secretion system protein VirB3 | virB3 |
| ELI21_RS30730 (ELI21_30735) | 301959..304325 | + | 2367 | WP_130671363 | VirB4 family type IV secretion system protein | virb4 |
| ELI21_RS30735 (ELI21_30740) | 304358..305023 | + | 666 | WP_245487114 | P-type DNA transfer protein VirB5 | virB5 |
| ELI21_RS30740 (ELI21_30745) | 305020..305253 | + | 234 | WP_130671365 | EexN family lipoprotein | - |
| ELI21_RS30745 (ELI21_30750) | 305257..306177 | + | 921 | WP_130671366 | type IV secretion system protein | virB6 |
| ELI21_RS30750 (ELI21_30755) | 306230..306514 | + | 285 | WP_130671367 | hypothetical protein | - |
| ELI21_RS30755 (ELI21_30760) | 306516..307187 | + | 672 | WP_130671368 | virB8 family protein | virB8 |
| ELI21_RS30760 (ELI21_30765) | 307184..308038 | + | 855 | WP_130671369 | P-type conjugative transfer protein VirB9 | virB9 |
| ELI21_RS30765 (ELI21_30770) | 308048..309220 | + | 1173 | WP_130671370 | type IV secretion system protein VirB10 | virB10 |
| ELI21_RS30770 (ELI21_30775) | 309227..310267 | + | 1041 | WP_130671371 | P-type DNA transfer ATPase VirB11 | virB11 |
| ELI21_RS35875 | 310410..310578 | - | 169 | Protein_295 | nucleotidyltransferase | - |
| ELI21_RS30775 (ELI21_30780) | 311052..311687 | + | 636 | WP_130671372 | invasion associated locus B family protein | - |
| ELI21_RS30780 (ELI21_30785) | 311700..313150 | + | 1451 | WP_130671373 | ShlB/FhaC/HecB family hemolysin secretion/activation protein | - |
Host bacterium
| ID | 2975 | GenBank | NZ_SIPU01000004 |
| Plasmid name | pSM152A_Rh03 | Incompatibility group | - |
| Plasmid size | 313150 bp | Coordinate of oriT [Strand] | 27756..27784 [+] |
| Host baterium | Rhizobium leguminosarum strain SM152A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |