Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102528 |
| Name | oriT_pSM168A_Rh08 |
| Organism | Rhizobium leguminosarum strain SM168A |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIQX01000004 (95211..95248 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_pSM168A_Rh08
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
GGATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file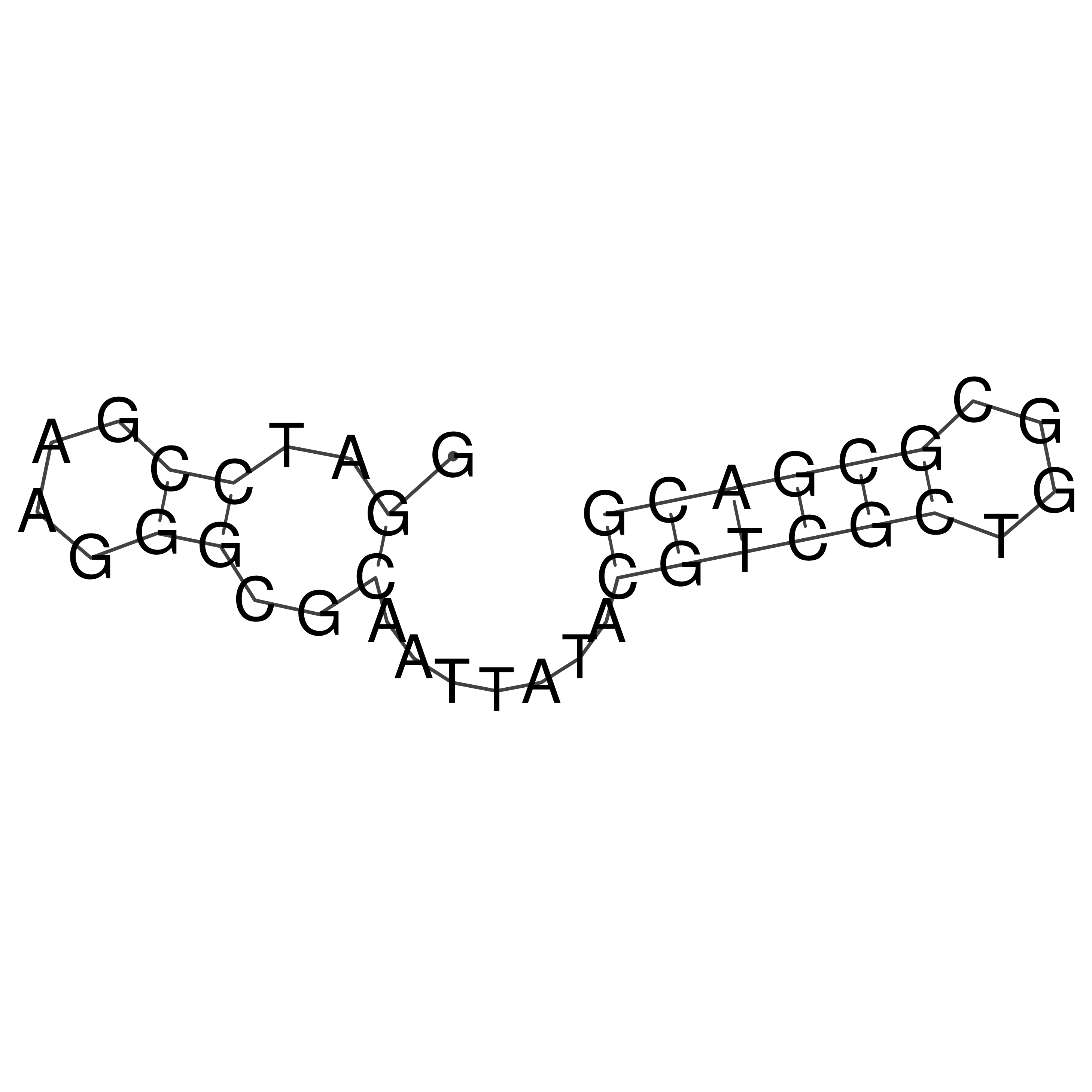
Relaxase
| ID | 1879 | GenBank | WP_130654295 |
| Name | traA_ELI50_RS32605_pSM168A_Rh08 |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 122889.15 Da Isoelectric Point: 9.8379
>WP_130654295.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLIEDGFGSKKVAILGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPTKYTTREM
IRIEAEMANHAIWLSARASHGVREAVLHATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNRLDDKTVIVLDEAGMVSSRQM
ALLVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLARVLEAASGRIVAEIGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYQ
KSALYRQALRFAEARGLQLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIRDTI
KEAKPMVSGITTFPKSLDQAVEGKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKAINVDAMLTDQSVAQS
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYTRKQGLLHEEFVIPADAPEWVRSMIADR
SVAGASEAFWNKVEGFEKRSDAQLAKDVTIALPLELTAEQNIALMRDFVAGHITAQGMVADWVYHDAPGN
PHVHLMTTLRPLIEDGFGSKKVAILGPDGKPIRNDAGKIVYELWAGSTEDFNAFRDGWFACQNKHLALAG
LDIRIDGRSFEKQGIDLEPTIHLGVGAKAIERKAEQSDHKSETSTPKLERIELQEERRSENARRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDVALFQSLMVRILQSPEALRLERERINFATGIRTPTKYTTREM
IRIEAEMANHAIWLSARASHGVREAVLHATFARHSRLSDEQRTAIEHVAGATRIAAIIGRAGAGKTTMMK
AAREAWETAGYRVVGGALAGKAAEGLEKEAGIVSRTLSSWELRWNQGRNRLDDKTVIVLDEAGMVSSRQM
ALLVEAVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRQQWMRDASLDLARGNVRKA
VDAYTAHGRMIGLRLKDEAVESLIAAWDRDYDPSKTSLILAHLRRDVRMLNDMARAKLVERGVVADGFAF
KTEDGTRMFATGDQIVFLKNEGSLGVKNGMLARVLEAASGRIVAEIGDGEHRRQVTVEQRFYNNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGSRSFAKSGGLIPILSRRNAKETSLDYQ
KSALYRQALRFAEARGLQLMNVARTIAHDRLQWAVRQKQKLADLGARLAAIGAALGLVRGSNKHSIRDTI
KEAKPMVSGITTFPKSLDQAVEGKLAADPGLKKQWEDVSTRFHLVYAQPEAAFKAINVDAMLTDQSVAQS
TLARIAGEPESFGALKGKTGLLASRGDKQDREKALANVPALARNLERYLRERAEAEFKHETEERAVRLKV
SVDIPALSPAAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFAKAVSERFGERTFLPLAAKDTDG
KTFETVTAGMTAGQKAEVRNAWDAMRAVQQMAAHERTTEALKQAETLRQTRSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1479 | GenBank | WP_130654280 |
| Name | t4cp2_ELI50_RS32520_pSM168A_Rh08 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91543.33 Da Isoelectric Point: 5.5133
>WP_130654280.1 conjugal transfer protein TrbE [Rhizobium leguminosarum]
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDIPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLESTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDRDREGLLEKAEAIRRLIQVEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGNSLLSLTLAAGADYYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMAIASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAAPLLK
MVALKPFRVTGPSFADLVPYAGLIDDGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSKDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEESRKKSYADTVLFVFKNAVRELEQYFANSLSIRRMETRETFERGGERIARYDELLQFARFCTTG
ESHPIRLPDIPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLESTRKKWQQKVRPFFDQIFQTQSRSVDQDAMTMVVETEDAIAQASSQLVAYGYYTPVVVL
FDRDREGLLEKAEAIRRLIQVEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTAFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGNSLLSLTLAAGADYYEIGGDNAEEGRALAFCPLSELKSDADRAWATEWIEMLVGLQGVTITPDHRN
AISRQIGLMAIASGRSLSDFVSGVQLREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVATATPKREYYVATPEGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSENGHDWPIHWLETRGVHDAAPLLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1480 | GenBank | WP_130654298 |
| Name | traG_ELI50_RS32620_pSM168A_Rh08 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 644 a.a. Molecular weight: 69354.56 Da Isoelectric Point: 9.2319
>WP_130654298.1 Ti-type conjugative transfer system protein TraG [Rhizobium leguminosarum]
MTINRIALVIVPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYGASAAIGILFLFASAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTKALNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLLPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGASG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
MTINRIALVIVPAALMILVVIGMTGIEQWLSGFGKTEAARLAWGRAGIALPYGASAAIGILFLFASAGSI
NIKQAGWGVLAGCSGTILIAATRETMRLAAFTKALNADKSVLAYLDPATAIGAGVALMCACFALRVVLIG
NAAFARAEPKRIQGKRALHGEADWMKLTEAAKLLPDAGGIVIGERYRVDKDNVGSQAFRADSAETWGASG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSNEVAPMVSAHRGGAGRDVFVLD
PRKPDIGFNVLDWVGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDG
HDQTLRQVRMNLSEPEPTLRKRLQDIYDNSGSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGKKFATNDIAAGNTDVFINIDLKTLETHSGLARVVIGSLLNAIYNRDGQMKGRALFLLDEVARLGY
MRIIETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHMVGDTPKPA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 75939..90075
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ELI50_RS32485 (ELI50_32485) | 71265..72479 | - | 1215 | WP_130654329 | plasmid replication protein RepC | - |
| ELI50_RS32490 (ELI50_32490) | 72670..73707 | - | 1038 | WP_130654275 | plasmid partitioning protein RepB | - |
| ELI50_RS32495 (ELI50_32495) | 73704..74927 | - | 1224 | WP_065276670 | plasmid partitioning protein RepA | - |
| ELI50_RS32500 (ELI50_32500) | 75304..75942 | + | 639 | WP_130654277 | acyl-homoserine-lactone synthase | - |
| ELI50_RS32505 (ELI50_32505) | 75939..76916 | + | 978 | WP_130654278 | P-type conjugative transfer ATPase TrbB | virB11 |
| ELI50_RS32510 (ELI50_32510) | 76906..77289 | + | 384 | WP_065276633 | TrbC/VirB2 family protein | virB2 |
| ELI50_RS32515 (ELI50_32515) | 77282..77581 | + | 300 | WP_130654279 | conjugal transfer protein TrbD | virB3 |
| ELI50_RS32520 (ELI50_32520) | 77592..80042 | + | 2451 | WP_130654280 | conjugal transfer protein TrbE | virb4 |
| ELI50_RS32525 (ELI50_32525) | 80020..80817 | + | 798 | WP_130654281 | P-type conjugative transfer protein TrbJ | virB5 |
| ELI50_RS32530 (ELI50_32530) | 80811..80987 | + | 177 | WP_130654282 | entry exclusion protein TrbK | - |
| ELI50_RS32535 (ELI50_32535) | 80992..82179 | + | 1188 | WP_130654283 | P-type conjugative transfer protein TrbL | virB6 |
| ELI50_RS32540 (ELI50_32540) | 82200..82862 | + | 663 | WP_130654284 | conjugal transfer protein TrbF | virB8 |
| ELI50_RS32545 (ELI50_32545) | 82880..83692 | + | 813 | WP_130654285 | P-type conjugative transfer protein TrbG | virB9 |
| ELI50_RS32550 (ELI50_32550) | 83696..84136 | + | 441 | WP_130654286 | conjugal transfer protein TrbH | - |
| ELI50_RS32555 (ELI50_32555) | 84148..85446 | + | 1299 | WP_130654287 | IncP-type conjugal transfer protein TrbI | virB10 |
| ELI50_RS32560 (ELI50_32560) | 85758..86462 | + | 705 | WP_130654288 | autoinducer binding domain-containing protein | - |
| ELI50_RS32565 (ELI50_32565) | 86466..86789 | - | 324 | WP_130654289 | transcriptional repressor TraM | - |
| ELI50_RS32570 (ELI50_32570) | 87012..87254 | + | 243 | WP_130654290 | helix-turn-helix transcriptional regulator | - |
| ELI50_RS37880 (ELI50_32575) | 87357..87788 | + | 432 | Protein_92 | hypothetical protein | - |
| ELI50_RS32580 (ELI50_32580) | 87879..88496 | + | 618 | WP_130654291 | type IV toxin-antitoxin system AbiEi family antitoxin domain-containing protein | - |
| ELI50_RS32585 (ELI50_32585) | 88489..89355 | + | 867 | Protein_94 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| ELI50_RS32590 (ELI50_32590) | 89458..90075 | - | 618 | WP_130654292 | TraH family protein | virB1 |
| ELI50_RS32595 (ELI50_32595) | 90092..91255 | - | 1164 | WP_130654293 | conjugal transfer protein TraB | - |
| ELI50_RS32600 (ELI50_32600) | 91245..91808 | - | 564 | WP_130654294 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 2972 | GenBank | NZ_SIQX01000004 |
| Plasmid name | pSM168A_Rh08 | Incompatibility group | - |
| Plasmid size | 153113 bp | Coordinate of oriT [Strand] | 95211..95248 [-] |
| Host baterium | Rhizobium leguminosarum strain SM168A |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD |
| Anti-CRISPR | - |