Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102520 |
| Name | oriT_pSM155C_Rh02 |
| Organism | Rhizobium leguminosarum strain SM155C |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_SIQF01000003 (389576..389604 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSM155C_Rh02
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file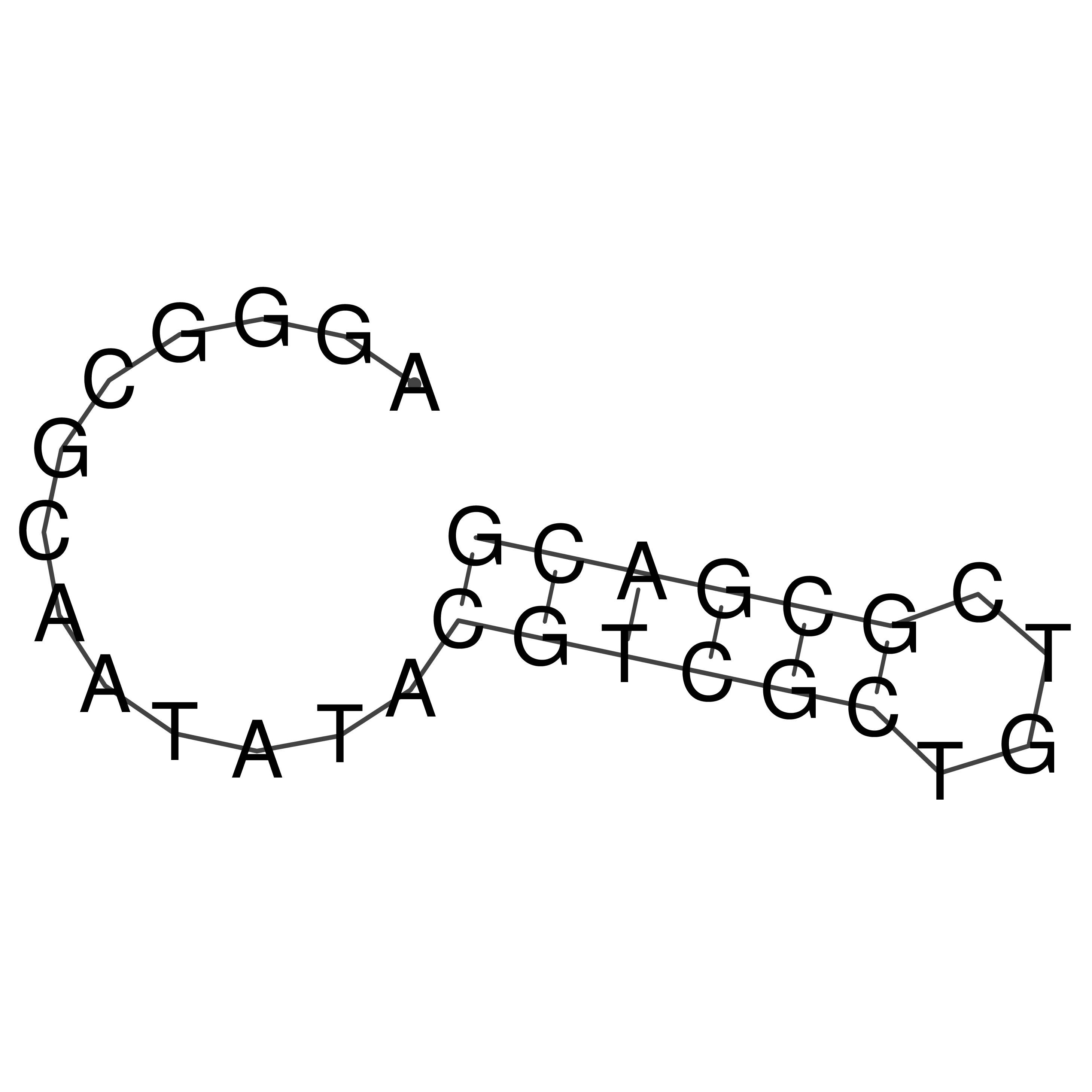
Relaxase
| ID | 1871 | GenBank | WP_130696272 |
| Name | traA_ELI32_RS30540_pSM155C_Rh02 |
UniProt ID | _ |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 170583.63 Da Isoelectric Point: 7.2557
>WP_130696272.1 Ti-type conjugative transfer relaxase TraA [Rhizobium leguminosarum]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQTPDWLRAAIDG
RSVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIGLVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTQEGFGRKKVAVTGENGAPLRVVTPDRPTGRIVYKLWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPALLLKQ
LSNERSTFDERDIAKALHRTVDDPLDFANIRARLMASDDLVMLKPQQVDTETGKAAEPAVFTTRDILRIE
YDMARSAQVLSERRGFAVSAESLAAAIENVETADPEKSFRLDPEQVDAVHHVTRDNGIAAVVGLAGAGKS
TLLAAARVAWESGGSRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWANGHETLHRGDVVVVDEAGMVS
SQQMARLLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQQWAREASRLFARGE
IEKGLDAYAQQGHLIEAGTRAETIDRIVADWSEARKRAIETAALKGNDGRLRGDELLVLAHTNQDVKRLN
EALRSVMTGEGALGENRSFQTERGGREFAAGDRIIFLENSRFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGSTVERTLVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHTAGVTGELVETGQAKFRPNDEDADDSPYADVKTEDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVKVQVAIVDEQTGQKRFEEREVERNVWTAKRIETAAERQQRIERESHRP
ELFKQLVERLSRSGAKTTTLDFESEAGYQAHADDFARRRGLYTHSQTAAGMEEGVSRRWVWIAEKREQLA
ELWERASVALGFAIERERRASYNEERIETQTVPDADAGRARYLLPSTTVFARSVEEDARLAQLASPAWKE
REAILRPLLEKIYRDPDAALVALNALASDKTIEPRRLADDLAVAPDRLGRVRGSDLIVDGRAARDERSIA
IAALTELIPLARAHATEFRRNAERFGVREQQRRAHMSLPIPALSKTAMARLVEIEAVRRQGGDDAYKTAF
ALATEDRSLVQEVKAVSEALTARFGWSAFTSNADALAERNMVERMPEDLASTRREKLTRLFEAVKRFADE
QHLAERRDRSKIVAGASADQGMEAGKENAAVLPMLAAVTEFRTPIDEEARSRALATPLYRQQRTALATVA
STIWRDPAGAVGKIEDLLAKGFAGDRIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKDAVQALPEAAA
CLRALGSAYVNMLAAERHAIAEERRRMAVAIPGLSKAAEEALMRLTAEAKNNGRKPNASAVSLDPGIRRE
FAAVSTALDARFGRNAILRDDKDLINRIPPVQRRAFEAMQERFKVLQQAVRIESSEQIISERRQRAVSRG
HGIDL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQTPDWLRAAIDG
RSVAAASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIGLVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTTLRPLTQEGFGRKKVAVTGENGAPLRVVTPDRPTGRIVYKLWAGDKETMKAWKVAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALSRRGAEMYFAPADLARRQEMADRLLADPALLLKQ
LSNERSTFDERDIAKALHRTVDDPLDFANIRARLMASDDLVMLKPQQVDTETGKAAEPAVFTTRDILRIE
YDMARSAQVLSERRGFAVSAESLAAAIENVETADPEKSFRLDPEQVDAVHHVTRDNGIAAVVGLAGAGKS
TLLAAARVAWESGGSRVIGAALAGKAAEGLEDSSGIRSRTLASWELIWANGHETLHRGDVVVVDEAGMVS
SQQMARLLKLVEQAGAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQQWAREASRLFARGE
IEKGLDAYAQQGHLIEAGTRAETIDRIVADWSEARKRAIETAALKGNDGRLRGDELLVLAHTNQDVKRLN
EALRSVMTGEGALGENRSFQTERGGREFAAGDRIIFLENSRFLEPRAPRLGPQYVKNGMLGTVVSTGDKR
GDTLLSVRLDNGRDVVISEDSYRNVDHGYAATIHKSQGSTVERTLVLATGMMDQHLTYVSMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHTAGVTGELVETGQAKFRPNDEDADDSPYADVKTEDGSVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVKVQVAIVDEQTGQKRFEEREVERNVWTAKRIETAAERQQRIERESHRP
ELFKQLVERLSRSGAKTTTLDFESEAGYQAHADDFARRRGLYTHSQTAAGMEEGVSRRWVWIAEKREQLA
ELWERASVALGFAIERERRASYNEERIETQTVPDADAGRARYLLPSTTVFARSVEEDARLAQLASPAWKE
REAILRPLLEKIYRDPDAALVALNALASDKTIEPRRLADDLAVAPDRLGRVRGSDLIVDGRAARDERSIA
IAALTELIPLARAHATEFRRNAERFGVREQQRRAHMSLPIPALSKTAMARLVEIEAVRRQGGDDAYKTAF
ALATEDRSLVQEVKAVSEALTARFGWSAFTSNADALAERNMVERMPEDLASTRREKLTRLFEAVKRFADE
QHLAERRDRSKIVAGASADQGMEAGKENAAVLPMLAAVTEFRTPIDEEARSRALATPLYRQQRTALATVA
STIWRDPAGAVGKIEDLLAKGFAGDRIAAAVTNDPAAYGALRGSDRLMDRMLAAGRERKDAVQALPEAAA
CLRALGSAYVNMLAAERHAIAEERRRMAVAIPGLSKAAEEALMRLTAEAKNNGRKPNASAVSLDPGIRRE
FAAVSTALDARFGRNAILRDDKDLINRIPPVQRRAFEAMQERFKVLQQAVRIESSEQIISERRQRAVSRG
HGIDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 2964 | GenBank | NZ_SIQF01000003 |
| Plasmid name | pSM155C_Rh02 | Incompatibility group | - |
| Plasmid size | 480486 bp | Coordinate of oriT [Strand] | 389576..389604 [+] |
| Host baterium | Rhizobium leguminosarum strain SM155C |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |