Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102516 |
| Name | oriT_DCY119|unnamed2 |
| Organism | Mesorhizobium sp. DCY119 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_QYZQ01000003 (122441..122469 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..20, 23..29 (CGTCGCG..CGCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_DCY119|unnamed2
AGGGCGCAATATACGTCGCGGTCGCGACG
AGGGCGCAATATACGTCGCGGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file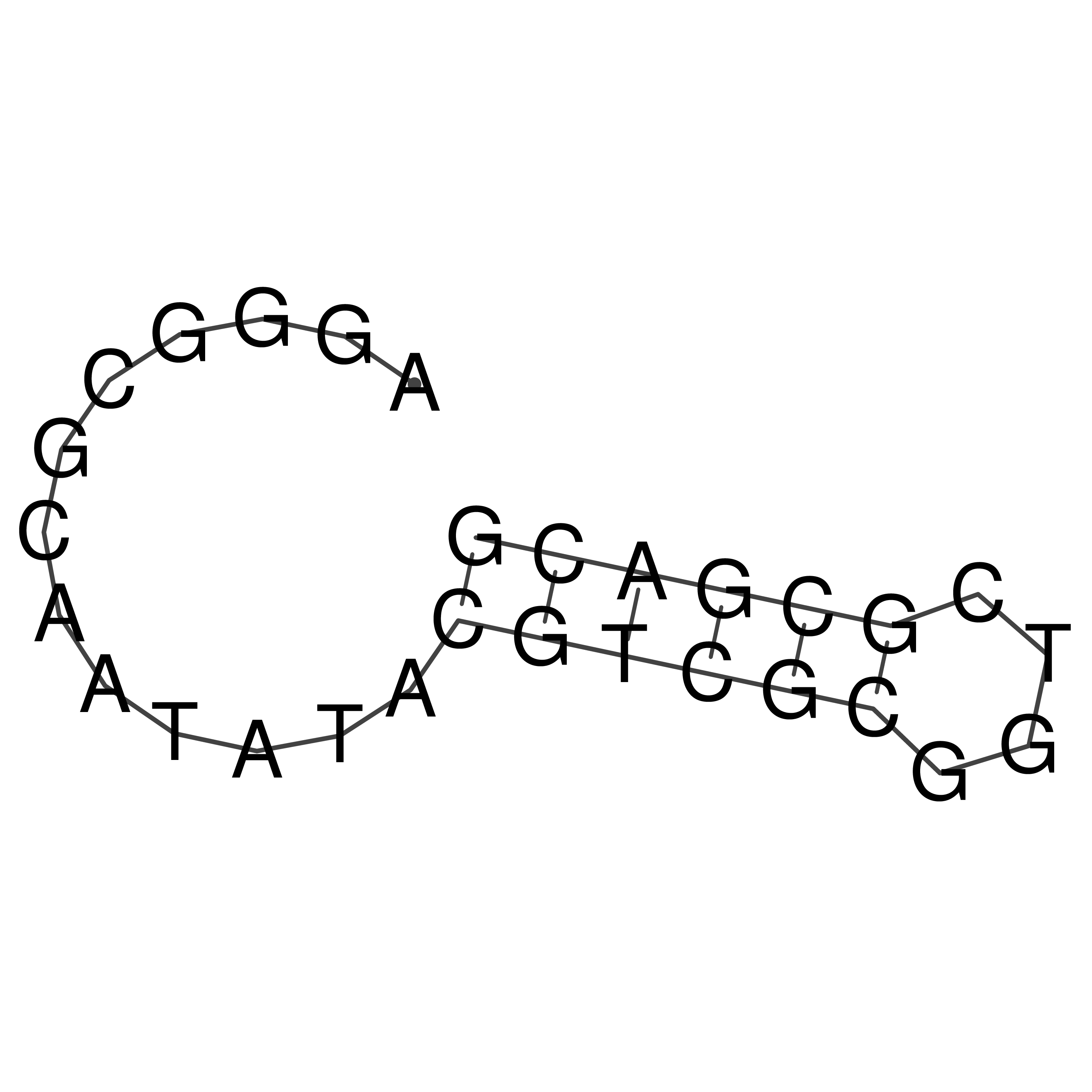
Relaxase
| ID | 1867 | GenBank | WP_119921868 |
| Name | traA_D3Y55_RS32235_DCY119|unnamed2 |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170670.43 Da Isoelectric Point: 8.6351
>WP_119921868.1 Ti-type conjugative transfer relaxase TraA [Mesorhizobium sp. DCY119]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAAELKHEELALPDQVPAWLRTAIDG
KTVAAASEALWNAVDAFETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGQKKVPVLGADGALLRVVTPDRPNGKIVYKLWAGDKETMKDWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLAEPELLLKQ
LANERSTFDEKDIAKALHRTVDDPADFANIRARLMASDDLVLLKQQQIDRQTGKVSEPAVFTTRDILRIE
YDMARSAEVLSQRKGFGIGEGKIAAAVRAIETADPEKSFMLDREQVEAVRHVAGDNGIAAVVGLAGAGKS
TLFAAARVAWESENHRVFGAALAGKAAEGLEDSSAIKSRTLASWEFAWANGRDLLDRGDVFVVDEAGMVS
SQQMARVLKIAEDAGAKVVLVGDAMQLQPIEAGAAFRAIVERIGSAELAGVRRQREEWAREASRLFARGK
VEEALDAYAREGRIFEAESRADVVERIVADWTDARRDLIRDTVDSEQPVRLRGDELLVLAHTNDDVKRLN
EALRKVMKDDGALTAARDFQTERGVREFAAGDRIIFLENARFLEPRARKLGPQYVKNGMRGTVVSTGDKR
GDPLLSVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRVRADL
YAAKEDFQPKLEWGRAARADHAAGVTGELVETGLAKFRPNDEDADESPYADMKTDNGAVQRLWGVSLPKA
LEEAGVSEGDTVTLRKDGVERVTVTVPIIDEKTGNKSFEERTVDRNVWTAELKETADVRRKRIERESHRP
ELFKQLVERLGRSGAKTTTLDFESEEGYRAHAQDFAQRRGIDTLAGIVAGMEEGVSGRRAWLAEKREQVA
KLWERASVALGFAIERERRVSYNEAQNETLRAEVPAGGKYLIPPTTMFARSVEEDARRAQLASERWKERD
AILRPVLQKIYRDPDGALARLNALASDARIEPRRLAEDLETKPQRLGRLRGSALLVDGRAARTERETAMA
ALTDLLPLARSHATEFRRQAERFGLREEQRRAHMSLSVPALSKPAMARLVEIEAVRERGGQDAYKTAFAY
AVADSLLVQEVKAVNDALTGRFGWSAFTQKADTVAERNTVERMPEDLAPDRREKLARLFAVVRRFAEEQH
LAERRDRSKTVAGASVEPGRETMPVLPMLAAVTEFKTPVEDEARERVRAVAHYGHNRAALVETAARLWRD
PAGAVDKIEALVVKGFAGDRIAAAVANDPAAYGALRGSDRIMDRMLAVGRERKDALQAVPEAASRVRSLG
ASYAAALDTETRTITEERRRMSVSIPGLSVSAEEALRDLSAAMKKKDAKLEVAAGALDPAIRSEFVTVSR
ALDDRFGRNAILRGEKDVINRVSPAQRRAFEVMQQRLQVLQQTVRMQASQEIVAERQRRVLDRARGVVR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGAAELKHEELALPDQVPAWLRTAIDG
KTVAAASEALWNAVDAFETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGQKKVPVLGADGALLRVVTPDRPNGKIVYKLWAGDKETMKDWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLAEPELLLKQ
LANERSTFDEKDIAKALHRTVDDPADFANIRARLMASDDLVLLKQQQIDRQTGKVSEPAVFTTRDILRIE
YDMARSAEVLSQRKGFGIGEGKIAAAVRAIETADPEKSFMLDREQVEAVRHVAGDNGIAAVVGLAGAGKS
TLFAAARVAWESENHRVFGAALAGKAAEGLEDSSAIKSRTLASWEFAWANGRDLLDRGDVFVVDEAGMVS
SQQMARVLKIAEDAGAKVVLVGDAMQLQPIEAGAAFRAIVERIGSAELAGVRRQREEWAREASRLFARGK
VEEALDAYAREGRIFEAESRADVVERIVADWTDARRDLIRDTVDSEQPVRLRGDELLVLAHTNDDVKRLN
EALRKVMKDDGALTAARDFQTERGVREFAAGDRIIFLENARFLEPRARKLGPQYVKNGMRGTVVSTGDKR
GDPLLSVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRVRADL
YAAKEDFQPKLEWGRAARADHAAGVTGELVETGLAKFRPNDEDADESPYADMKTDNGAVQRLWGVSLPKA
LEEAGVSEGDTVTLRKDGVERVTVTVPIIDEKTGNKSFEERTVDRNVWTAELKETADVRRKRIERESHRP
ELFKQLVERLGRSGAKTTTLDFESEEGYRAHAQDFAQRRGIDTLAGIVAGMEEGVSGRRAWLAEKREQVA
KLWERASVALGFAIERERRVSYNEAQNETLRAEVPAGGKYLIPPTTMFARSVEEDARRAQLASERWKERD
AILRPVLQKIYRDPDGALARLNALASDARIEPRRLAEDLETKPQRLGRLRGSALLVDGRAARTERETAMA
ALTDLLPLARSHATEFRRQAERFGLREEQRRAHMSLSVPALSKPAMARLVEIEAVRERGGQDAYKTAFAY
AVADSLLVQEVKAVNDALTGRFGWSAFTQKADTVAERNTVERMPEDLAPDRREKLARLFAVVRRFAEEQH
LAERRDRSKTVAGASVEPGRETMPVLPMLAAVTEFKTPVEDEARERVRAVAHYGHNRAALVETAARLWRD
PAGAVDKIEALVVKGFAGDRIAAAVANDPAAYGALRGSDRIMDRMLAVGRERKDALQAVPEAASRVRSLG
ASYAAALDTETRTITEERRRMSVSIPGLSVSAEEALRDLSAAMKKKDAKLEVAAGALDPAIRSEFVTVSR
ALDDRFGRNAILRGEKDVINRVSPAQRRAFEVMQQRLQVLQQTVRMQASQEIVAERQRRVLDRARGVVR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1464 | GenBank | WP_119921871 |
| Name | traG_D3Y55_RS32250_DCY119|unnamed2 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 70324.12 Da Isoelectric Point: 9.2774
>WP_119921871.1 Ti-type conjugative transfer system protein TraG [Mesorhizobium sp. DCY119]
MTIRAKPHPSLLLILVPITVTAFATYVVGWRWPEIAAGMTGKTQYWFLRASPVPALLFGPLAGLLAAWAL
PLHRRRPIAMATLICFLSVTGFYALREFGRLSPPVEGGAVTWNQALSFIDMMAVVGAIVGFMATAVSARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVVGERYRVDKEIVHELPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQNSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFRPSDLVGGKIDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLLYQSVGQLERHFGRDGAVSWIDGCAFASYAAIKAVETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESLNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAVYFRRKDMNAEAK
VNRFVKP
MTIRAKPHPSLLLILVPITVTAFATYVVGWRWPEIAAGMTGKTQYWFLRASPVPALLFGPLAGLLAAWAL
PLHRRRPIAMATLICFLSVTGFYALREFGRLSPPVEGGAVTWNQALSFIDMMAVVGAIVGFMATAVSARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVVGERYRVDKEIVHELPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQNSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFRPSDLVGGKIDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLLYQSVGQLERHFGRDGAVSWIDGCAFASYAAIKAVETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESLNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAVYFRRKDMNAEAK
VNRFVKP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 8648..18312
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| D3Y55_RS31670 (D3Y55_31680) | 4600..5595 | - | 996 | WP_119921774 | plasmid partitioning protein RepB | - |
| D3Y55_RS31675 (D3Y55_31685) | 5680..6870 | - | 1191 | WP_119921775 | plasmid partitioning protein RepA | - |
| D3Y55_RS31680 (D3Y55_31690) | 7247..7633 | + | 387 | WP_119921776 | hypothetical protein | - |
| D3Y55_RS31685 (D3Y55_31695) | 7650..8012 | - | 363 | WP_119921777 | transcriptional regulator | - |
| D3Y55_RS31690 (D3Y55_31700) | 8106..8645 | + | 540 | WP_119921778 | hypothetical protein | - |
| D3Y55_RS31695 (D3Y55_31705) | 8648..9334 | + | 687 | WP_119921779 | lytic transglycosylase domain-containing protein | virB1 |
| D3Y55_RS31700 (D3Y55_31710) | 9331..9630 | + | 300 | WP_119921780 | TrbC/VirB2 family protein | virB2 |
| D3Y55_RS31705 (D3Y55_31715) | 9633..9974 | + | 342 | WP_119921781 | type IV secretion system protein VirB3 | virB3 |
| D3Y55_RS31710 (D3Y55_31720) | 9967..12348 | + | 2382 | WP_119921782 | VirB4 family type IV secretion system protein | virb4 |
| D3Y55_RS31715 (D3Y55_31725) | 12348..13046 | + | 699 | WP_119921783 | P-type DNA transfer protein VirB5 | virB5 |
| D3Y55_RS31720 (D3Y55_31730) | 13043..13276 | + | 234 | WP_119921784 | EexN family lipoprotein | - |
| D3Y55_RS31725 (D3Y55_31735) | 13281..14216 | + | 936 | WP_119921785 | type IV secretion system protein | virB6 |
| D3Y55_RS33280 (D3Y55_31740) | 14254..14520 | + | 267 | WP_119921786 | hypothetical protein | - |
| D3Y55_RS31735 (D3Y55_31745) | 14524..15195 | + | 672 | WP_119921787 | virB8 family protein | virB8 |
| D3Y55_RS31740 (D3Y55_31750) | 15192..16046 | + | 855 | WP_165867741 | P-type conjugative transfer protein VirB9 | virB9 |
| D3Y55_RS31745 (D3Y55_31755) | 16061..17233 | + | 1173 | WP_119921788 | type IV secretion system protein VirB10 | virB10 |
| D3Y55_RS31750 (D3Y55_31760) | 17242..18312 | + | 1071 | WP_119921789 | P-type DNA transfer ATPase VirB11 | virB11 |
| D3Y55_RS31755 (D3Y55_31765) | 18811..19011 | + | 201 | WP_119921790 | site-specific integrase | - |
| D3Y55_RS31760 (D3Y55_31770) | 19035..19307 | + | 273 | WP_119921791 | transposase | - |
| D3Y55_RS33675 (D3Y55_31775) | 19320..19445 | + | 126 | Protein_23 | integrase | - |
| D3Y55_RS31770 (D3Y55_31780) | 19496..20224 | - | 729 | WP_119921792 | amino acid ABC transporter ATP-binding protein | - |
| D3Y55_RS31775 (D3Y55_31785) | 20249..20941 | - | 693 | WP_119921793 | ABC transporter permease subunit | - |
| D3Y55_RS31780 (D3Y55_31790) | 20968..21687 | - | 720 | WP_119921794 | amino acid ABC transporter permease | - |
| D3Y55_RS31785 (D3Y55_31795) | 21735..22655 | - | 921 | WP_245443980 | amino acid ABC transporter substrate-binding protein | - |
Host bacterium
| ID | 2960 | GenBank | NZ_QYZQ01000003 |
| Plasmid name | DCY119|unnamed2 | Incompatibility group | - |
| Plasmid size | 170544 bp | Coordinate of oriT [Strand] | 122441..122469 [-] |
| Host baterium | Mesorhizobium sp. DCY119 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | bhpD |
| Symbiosis gene | - |
| Anti-CRISPR | - |