Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102508 |
| Name | oriT1_BMH-17-0036|unnamed2 |
| Organism | Escherichia coli strain BMH-17-0036 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAHMAA010000006 (21518..21597 [+], 80 nt) |
| oriT length | 80 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 80 nt
GGGTGTCGGGGCAAAGCCCTGACCAGGTGGTAATCGTATAGCCGTGCGTGCGCGGTTATACGATTACACATCCTGTCCCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file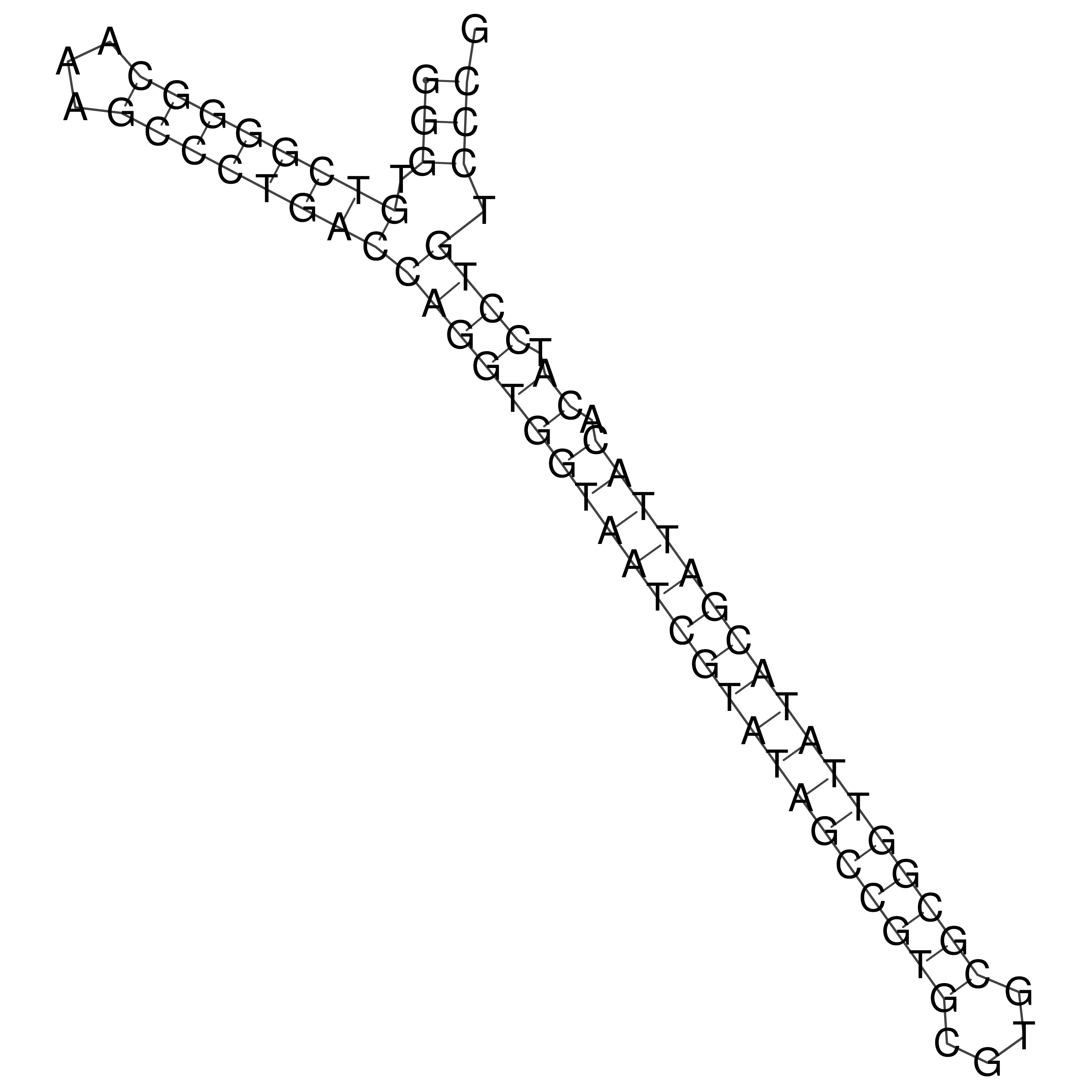
Relaxase
| ID | 1862 | GenBank | WP_042968735 |
| Name | traI_KQ660_RS28305_BMH-17-0036|unnamed2 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191689.42 Da Isoelectric Point: 5.6858
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMLT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQVVTQAIAGLSERKVQFTYTDVLVRTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYLW
QGGEQRPATIISEPDRNVRYARLAGDFAVSVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKIPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPATLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTSFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVKMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDVREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVMAVSGDSVTLSDGQQTRVIRPGQEQAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAISKAVQKGTAHDVL
EPGADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAISRETERRADEIVRKMAEEKPDQP
DGKTEQAVRAIAGQEQDRASSSERETALPESVLRESQRVQDAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 771 | GenBank | WP_032142519 |
| Name | WP_032142519_BMH-17-0036|unnamed2 |
UniProt ID | _ |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9106.17 Da Isoelectric Point: 10.3319
MRRRNARGGTSRTVSVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRYPDFYNEEIFREVTEERES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 772 | GenBank | WP_042968749 |
| Name | WP_042968749_BMH-17-0036|unnamed2 |
UniProt ID | _ |
| Length | 128 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 128 a.a. Molecular weight: 14643.71 Da Isoelectric Point: 4.7197
MAKVQAYVSDEIVYKINEIVERRRAEGAKSTDVSFSSISTMLLELGLRVYEAQMERKESSFNQTEFNKVL
LECVVKTQSSVAKILGIESLSPHIAGNPKFEYANMVEDIREKVSIEMDRFFPKIDNEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1460 | GenBank | WP_062872717 |
| Name | traD_KQ660_RS28310_BMH-17-0036|unnamed2 |
UniProt ID | _ |
| Length | 720 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 720 a.a. Molecular weight: 81893.41 Da Isoelectric Point: 5.1711
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVSTGKSEVIRRLANYAR
KRGDMVVIYDRSCEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNVANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTYLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHSGESFTIRDWMRGVREDKQNGWLFISSNADTHASLKPVVSMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVLNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKEMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARAKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPEVASGEDVTQAEQPQQPQQPQQPVS
PAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMTAYEAWQQENHPDIQQQMQRRE
EVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1461 | GenBank | WP_042968744 |
| Name | traC_KQ660_RS28425_BMH-17-0036|unnamed2 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99292.95 Da Isoelectric Point: 5.6251
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDSTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAE
TLFPLPEGLNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGAYITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERIRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLFPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRQEGL
SIHEAVWQLAWKKSGPEMASLESWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 40773..70143
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KQ660_RS28310 (KQ660_28200) | 40773..42935 | - | 2163 | WP_062872717 | type IV conjugative transfer system coupling protein TraD | virb4 |
| KQ660_RS28315 (KQ660_28205) | 42991..43722 | - | 732 | WP_000199899 | hypothetical protein | - |
| KQ660_RS28320 (KQ660_28210) | 43890..44150 | - | 261 | WP_001077900 | hypothetical protein | - |
| KQ660_RS28325 (KQ660_28215) | 44210..44941 | - | 732 | WP_000782449 | conjugal transfer complement resistance protein TraT | - |
| KQ660_RS28330 (KQ660_28220) | 44990..45469 | - | 480 | WP_021573915 | surface exclusion inner membrane protein | - |
| KQ660_RS28335 (KQ660_28225) | 45491..48334 | - | 2844 | WP_001007003 | conjugal transfer mating-pair stabilization protein TraG | traG |
| KQ660_RS28340 (KQ660_28230) | 48331..49704 | - | 1374 | WP_000944326 | conjugal transfer pilus assembly protein TraH | traH |
| KQ660_RS28345 (KQ660_28235) | 49691..50083 | - | 393 | WP_000662210 | F-type conjugal transfer protein TrbF | - |
| KQ660_RS28350 (KQ660_28240) | 50037..50351 | - | 315 | WP_001244902 | P-type conjugative transfer protein TrbJ | - |
| KQ660_RS28355 (KQ660_28245) | 50341..50895 | - | 555 | WP_000059845 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| KQ660_RS28360 (KQ660_28250) | 50882..51163 | - | 282 | WP_000049684 | type-F conjugative transfer system pilin chaperone TraQ | - |
| KQ660_RS28365 (KQ660_28255) | 51229..51630 | - | 402 | WP_225394595 | hypothetical protein | - |
| KQ660_RS28370 (KQ660_28260) | 51761..52126 | - | 366 | WP_001009441 | hypothetical protein | - |
| KQ660_RS28375 (KQ660_28265) | 52140..52883 | - | 744 | WP_001030408 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| KQ660_RS28380 (KQ660_28270) | 52876..53133 | - | 258 | WP_000864350 | conjugal transfer protein TrbE | - |
| KQ660_RS28385 (KQ660_28275) | 53147..54997 | - | 1851 | WP_216691226 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| KQ660_RS28390 (KQ660_28280) | 54994..55416 | - | 423 | WP_001229311 | hypothetical protein | - |
| KQ660_RS28395 (KQ660_28285) | 55442..55813 | - | 372 | WP_000056336 | hypothetical protein | - |
| KQ660_RS28400 (KQ660_28290) | 55810..56448 | - | 639 | WP_000087538 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| KQ660_RS28405 (KQ660_28295) | 56475..56996 | - | 522 | WP_000505570 | hypothetical protein | - |
| KQ660_RS28410 (KQ660_28300) | 57401..58393 | - | 993 | WP_032210964 | conjugal transfer pilus assembly protein TraU | traU |
| KQ660_RS28415 (KQ660_28305) | 58390..59022 | - | 633 | WP_000877292 | type-F conjugative transfer system protein TraW | traW |
| KQ660_RS28420 (KQ660_28310) | 59019..59405 | - | 387 | WP_000214100 | type-F conjugative transfer system protein TrbI | - |
| KQ660_RS28425 (KQ660_28315) | 59402..62029 | - | 2628 | WP_042968744 | type IV secretion system protein TraC | virb4 |
| KQ660_RS28430 (KQ660_28320) | 62189..62410 | - | 222 | WP_001278978 | conjugal transfer protein TraR | - |
| KQ660_RS28435 (KQ660_28325) | 62545..63060 | - | 516 | WP_042968745 | type IV conjugative transfer system lipoprotein TraV | traV |
| KQ660_RS28440 (KQ660_28330) | 63057..63308 | - | 252 | WP_001038336 | conjugal transfer protein TrbG | - |
| KQ660_RS28445 (KQ660_28335) | 63301..63621 | - | 321 | WP_001057265 | conjugal transfer protein TrbD | virb4 |
| KQ660_RS28450 (KQ660_28340) | 63608..64195 | - | 588 | WP_000002896 | conjugal transfer pilus-stabilizing protein TraP | - |
| KQ660_RS28455 (KQ660_28345) | 64185..65615 | - | 1431 | WP_000146622 | F-type conjugal transfer pilus assembly protein TraB | traB |
| KQ660_RS28460 (KQ660_28350) | 65615..66343 | - | 729 | WP_021573902 | type-F conjugative transfer system secretin TraK | traK |
| KQ660_RS28465 (KQ660_28355) | 66330..66896 | - | 567 | WP_000399759 | type IV conjugative transfer system protein TraE | traE |
| KQ660_RS28470 (KQ660_28360) | 66918..67229 | - | 312 | WP_000012108 | type IV conjugative transfer system protein TraL | traL |
| KQ660_RS28475 (KQ660_28365) | 67244..67597 | - | 354 | WP_001098217 | type IV conjugative transfer system pilin TraA | - |
| KQ660_RS28480 (KQ660_28370) | 67632..67859 | - | 228 | WP_032142519 | conjugal transfer relaxosome protein TraY | - |
| KQ660_RS28485 (KQ660_28375) | 67947..68636 | - | 690 | WP_042968747 | PAS domain-containing protein | - |
| KQ660_RS28490 (KQ660_28380) | 68825..69211 | - | 387 | WP_042968749 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| KQ660_RS28495 (KQ660_28385) | 69496..70143 | + | 648 | WP_077239113 | transglycosylase SLT domain-containing protein | virB1 |
| KQ660_RS28500 (KQ660_28390) | 70439..71260 | - | 822 | WP_001234499 | DUF932 domain-containing protein | - |
| KQ660_RS28505 (KQ660_28395) | 71379..71666 | - | 288 | WP_000107543 | hypothetical protein | - |
| KQ660_RS28510 (KQ660_28400) | 71822..72124 | - | 303 | WP_001272235 | hypothetical protein | - |
| KQ660_RS28515 | 72424..72597 | + | 174 | Protein_79 | hypothetical protein | - |
| KQ660_RS28520 (KQ660_28405) | 73051..73173 | - | 123 | WP_223200913 | Hok/Gef family protein | - |
| KQ660_RS28525 | 73118..73270 | - | 153 | Protein_81 | DUF5431 family protein | - |
| KQ660_RS28530 (KQ660_28410) | 73420..73734 | - | 315 | WP_000872086 | hypothetical protein | - |
| KQ660_RS28535 (KQ660_28415) | 73731..74450 | - | 720 | WP_032210982 | plasmid SOS inhibition protein A | - |
| KQ660_RS28540 (KQ660_28420) | 74447..74881 | - | 435 | WP_000845875 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 2952 | GenBank | NZ_JAHMAA010000006 |
| Plasmid name | BMH-17-0036|unnamed2 | Incompatibility group | IncFIB |
| Plasmid size | 113498 bp | Coordinate of oriT [Strand] | 21518..21597 [+]; 69448..69571 [-] |
| Host baterium | Escherichia coli strain BMH-17-0036 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hlyD, hlyB, hlyA, hlyC |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |