Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102506 |
| Name | oriT_L4M4-CTX03|unnamed |
| Organism | Escherichia coli strain L4M4-CTX03 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAQIZC010000002 (51008..51131 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 88..93, 105..110 (ATAAAT..ATTTAT) 39..45, 50..56 (GCAAAAA..TTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAATTTTTGCTGATTTGTTTTTTGAATCATTAGCTTATGTTATAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file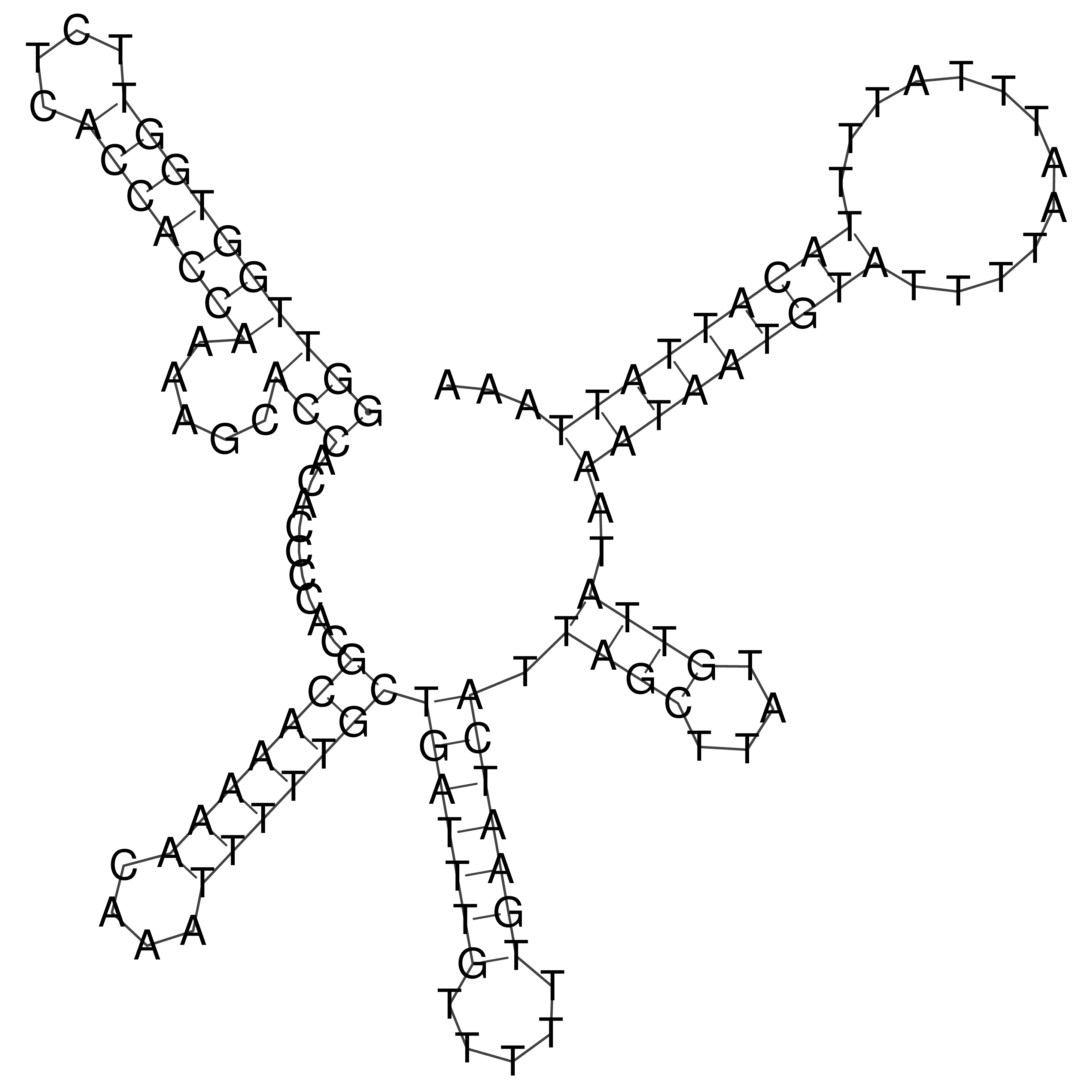
Relaxase
| ID | 1861 | GenBank | WP_047667298 |
| Name | traI_PIA89_RS02020_L4M4-CTX03|unnamed |
UniProt ID | A0A3Q8VPW8 |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191897.75 Da Isoelectric Point: 6.0145
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREKLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNIYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPRLRVSGGDRLQVASVSEDAMTVVVPGRAEPATLPVADSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISLQKTGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPASERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGETFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDALALVDSVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAQRLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVRDIAGQERDRSAISERETALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 769 | GenBank | WP_000124979 |
| Name | WP_000124979_L4M4-CTX03|unnamed |
UniProt ID | Q5JBL6 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14456.50 Da Isoelectric Point: 5.2973
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECAVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDGE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q5JBL6 |
| ID | 770 | GenBank | WP_001254385 |
| Name | WP_001254385_L4M4-CTX03|unnamed |
UniProt ID | A0A1Q9L2K4 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8975.08 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVAEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1Q9L2K4 |
T4CP
| ID | 1456 | GenBank | WP_000069740 |
| Name | traC_PIA89_RS01925_L4M4-CTX03|unnamed |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99356.04 Da Isoelectric Point: 6.0797
MSNNPLDAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAA
ATQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKKNKARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1457 | GenBank | WP_097416344 |
| Name | traD_PIA89_RS02015_L4M4-CTX03|unnamed |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 83583.22 Da Isoelectric Point: 5.0504
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQTRPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQ
QENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1458 | GenBank | WP_001332879 |
| Name | yjgR_PIA89_RS02110_L4M4-CTX03|unnamed |
UniProt ID | _ |
| Length | 500 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 500 a.a. Molecular weight: 54209.22 Da Isoelectric Point: 6.1946
MSEPLLIARTPDTELFLLPGMANRHGLITGATGTGKTVTLQKLAESLSEIGVPVFMADVKGDLTGIAQAG
TASEKLLARLKNIGVNDWQPHANPVVVWDIFGEKGHPVRATVSDLGPLLLARLLNLNDVQSGVLNIIFRI
ADDQGLLLLDFKDLRAITQYIGDNAKSFQNQYGNISSASVGAIQRGLLSLEQQGAAHFFGEPMLDIKDLM
RTDANGKGVINILSAEKLYQMPKLYAASLLWMISELYEQLPEAGDLEKPKLVFFFDEAHLLFNDAPQVLL
DKIEQVIRLIRSKGVGVWFVSQNPSDIPDNVLGQLGNRVQHALRAFTPKDQKAVKAAAQTMRANPAFDTE
KAIQELGTGEALISFLDAKGSPSVVERAMVIAPCSRMGPVTEDERNGLINHSPVYGKYEDDVDRESAYEM
LQKGVQASIEQQNNPPAKGKEIAVDDGILGGLKDILFGTTGPRGGKKDGVVQTMAKSAARQVTNQIVRGM
LGSLLGGRRR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 50436..79007
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PIA89_RS01780 (PIA89_01780) | 46402..46836 | + | 435 | WP_000845894 | conjugation system SOS inhibitor PsiB | - |
| PIA89_RS01785 (PIA89_01785) | 46833..47595 | + | 763 | Protein_67 | plasmid SOS inhibition protein A | - |
| PIA89_RS01790 (PIA89_01790) | 47564..47752 | - | 189 | WP_001299721 | hypothetical protein | - |
| PIA89_RS01795 (PIA89_01795) | 47774..47923 | + | 150 | Protein_69 | plasmid maintenance protein Mok | - |
| PIA89_RS01800 (PIA89_01800) | 47865..47990 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| PIA89_RS01805 (PIA89_01805) | 48210..48440 | + | 231 | WP_001426396 | hypothetical protein | - |
| PIA89_RS01810 (PIA89_01810) | 48438..48611 | - | 174 | Protein_72 | hypothetical protein | - |
| PIA89_RS01815 (PIA89_01815) | 48681..48887 | + | 207 | WP_000547968 | hypothetical protein | - |
| PIA89_RS01820 (PIA89_01820) | 48912..49199 | + | 288 | WP_000107535 | hypothetical protein | - |
| PIA89_RS01825 (PIA89_01825) | 49318..50139 | + | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| PIA89_RS01830 (PIA89_01830) | 50436..51083 | - | 648 | WP_139119334 | transglycosylase SLT domain-containing protein | virB1 |
| PIA89_RS01835 (PIA89_01835) | 51360..51743 | + | 384 | WP_000124979 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| PIA89_RS01840 (PIA89_01840) | 51934..52620 | + | 687 | WP_062880639 | PAS domain-containing protein | - |
| PIA89_RS01845 (PIA89_01845) | 52714..52941 | + | 228 | WP_001254385 | conjugal transfer relaxosome protein TraY | - |
| PIA89_RS01850 (PIA89_01850) | 52975..53334 | + | 360 | WP_000340272 | type IV conjugative transfer system pilin TraA | - |
| PIA89_RS01855 (PIA89_01855) | 53349..53660 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| PIA89_RS01860 (PIA89_01860) | 53682..54248 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| PIA89_RS01865 (PIA89_01865) | 54235..54963 | + | 729 | WP_001230786 | type-F conjugative transfer system secretin TraK | traK |
| PIA89_RS01870 (PIA89_01870) | 54963..56390 | + | 1428 | WP_000146655 | F-type conjugal transfer pilus assembly protein TraB | traB |
| PIA89_RS01875 (PIA89_01875) | 56380..56964 | + | 585 | WP_001299233 | conjugal transfer pilus-stabilizing protein TraP | - |
| PIA89_RS01880 (PIA89_01880) | 56951..57271 | + | 321 | WP_001057287 | conjugal transfer protein TrbD | virb4 |
| PIA89_RS01885 (PIA89_01885) | 57264..57515 | + | 252 | WP_001038338 | conjugal transfer protein TrbG | - |
| PIA89_RS01890 (PIA89_01890) | 57512..58027 | + | 516 | WP_000809833 | type IV conjugative transfer system lipoprotein TraV | traV |
| PIA89_RS01895 (PIA89_01895) | 58162..58383 | + | 222 | WP_001278696 | conjugal transfer protein TraR | - |
| PIA89_RS01900 (PIA89_01900) | 58376..58789 | + | 414 | WP_000549581 | hypothetical protein | - |
| PIA89_RS01905 (PIA89_01905) | 58782..59255 | + | 474 | WP_000549572 | hypothetical protein | - |
| PIA89_RS01910 (PIA89_01910) | 59334..59550 | + | 217 | Protein_92 | hypothetical protein | - |
| PIA89_RS01915 (PIA89_01915) | 59551..59862 | + | 312 | WP_000141580 | hypothetical protein | - |
| PIA89_RS01920 (PIA89_01920) | 59932..60294 | + | 363 | WP_000835374 | hypothetical protein | - |
| PIA89_RS01925 (PIA89_01925) | 60420..63050 | + | 2631 | WP_000069740 | type IV secretion system protein TraC | virb4 |
| PIA89_RS01930 (PIA89_01930) | 63047..63433 | + | 387 | WP_000214084 | type-F conjugative transfer system protein TrbI | - |
| PIA89_RS01935 (PIA89_01935) | 63430..64062 | + | 633 | WP_001203746 | type-F conjugative transfer system protein TraW | traW |
| PIA89_RS01940 (PIA89_01940) | 64059..65051 | + | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| PIA89_RS01945 (PIA89_01945) | 65077..65538 | + | 462 | WP_000277845 | hypothetical protein | - |
| PIA89_RS01950 (PIA89_01950) | 65568..66020 | + | 453 | WP_000069427 | hypothetical protein | - |
| PIA89_RS01955 (PIA89_01955) | 66048..66686 | + | 639 | WP_001104248 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| PIA89_RS01960 (PIA89_01960) | 66683..68491 | + | 1809 | WP_000821823 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| PIA89_RS01965 (PIA89_01965) | 68518..68775 | + | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| PIA89_RS01970 (PIA89_01970) | 68768..69511 | + | 744 | WP_001030376 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| PIA89_RS01975 (PIA89_01975) | 69527..69874 | + | 348 | WP_001287893 | conjugal transfer protein TrbA | - |
| PIA89_RS01980 (PIA89_01980) | 69993..70277 | + | 285 | WP_000624110 | type-F conjugative transfer system pilin chaperone TraQ | - |
| PIA89_RS01985 (PIA89_01985) | 70264..70809 | + | 546 | WP_000052614 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| PIA89_RS01990 (PIA89_01990) | 70739..71101 | + | 363 | WP_023181029 | P-type conjugative transfer protein TrbJ | - |
| PIA89_RS01995 (PIA89_01995) | 71101..72474 | + | 1374 | WP_021530364 | conjugal transfer pilus assembly protein TraH | traH |
| PIA89_RS02000 (PIA89_02000) | 72471..75296 | + | 2826 | WP_001007030 | conjugal transfer mating-pair stabilization protein TraG | traG |
| PIA89_RS02005 (PIA89_02005) | 75293..75802 | + | 510 | WP_000628099 | conjugal transfer entry exclusion protein TraS | - |
| PIA89_RS02010 (PIA89_02010) | 75816..76547 | + | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| PIA89_RS02015 (PIA89_02015) | 76800..79007 | + | 2208 | WP_097416344 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2950 | GenBank | NZ_JAQIZC010000002 |
| Plasmid name | L4M4-CTX03|unnamed | Incompatibility group | IncFII |
| Plasmid size | 233201 bp | Coordinate of oriT [Strand] | 51008..51131 [+] |
| Host baterium | Escherichia coli strain L4M4-CTX03 |
Cargo genes
| Drug resistance gene | sul3, ant(3'')-Ia, cmlA1, aadA2 |
| Virulence gene | htpB |
| Metal resistance gene | merR, merT, merP, merA, merD, merE, mgtA, cutA |
| Degradation gene | bhpD |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |