Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102488 |
| Name | oriT_pRphCh2410d |
| Organism | Rhizobium phaseoli Ch24-10 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AHJU02000028 (972323..972351 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRphCh2410d
AGGGCGCAATATACGTCGCTGTCGCGACG
AGGGCGCAATATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file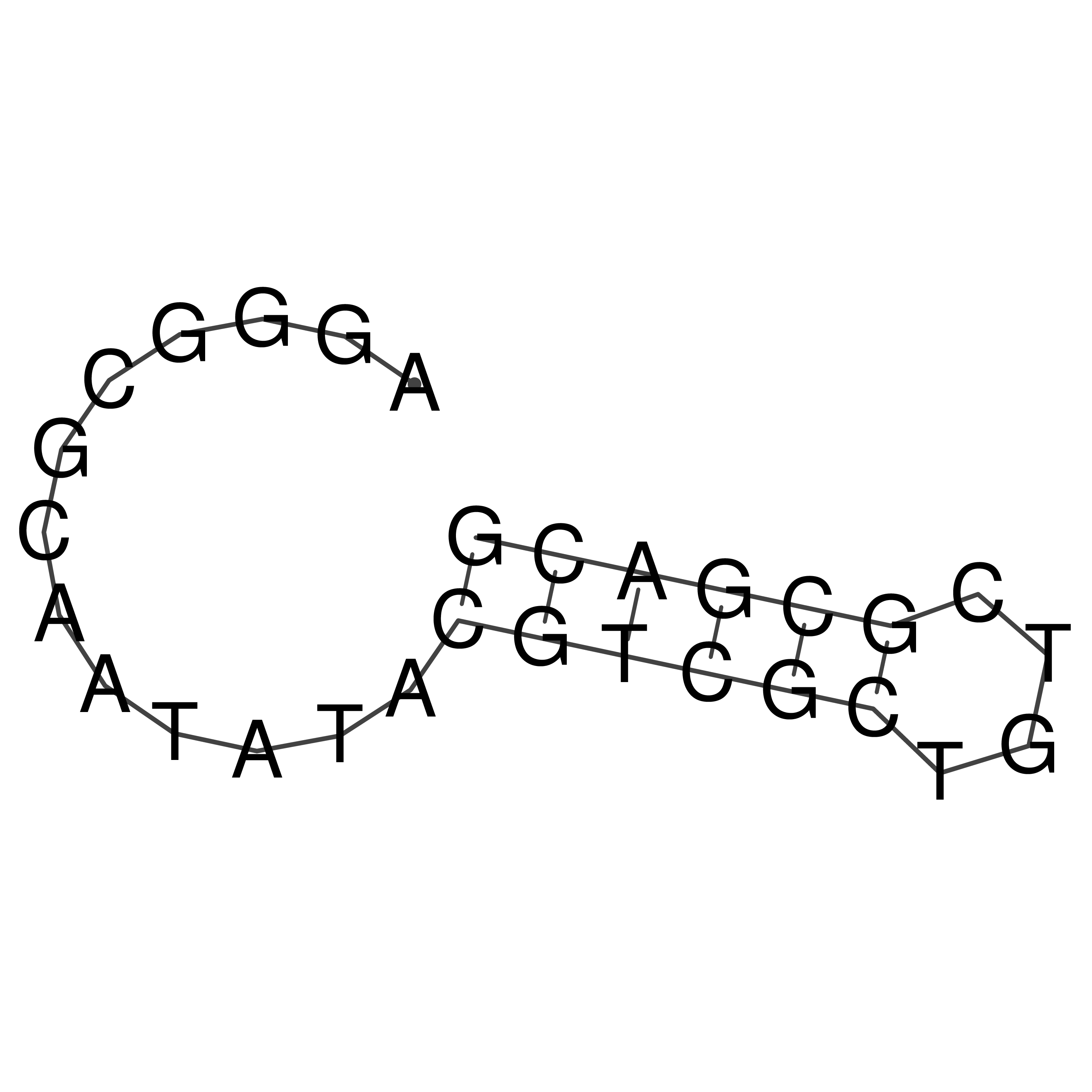
Relaxase
| ID | 1852 | GenBank | WP_046977228 |
| Name | traA_RPHASCH2410_RS29705_pRphCh2410d |
UniProt ID | _ |
| Length | 1543 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1543 a.a. Molecular weight: 170684.10 Da Isoelectric Point: 7.1240
>WP_046977228.1 Ti-type conjugative transfer relaxase TraA [Rhizobium phaseoli]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHDIRLDGRSYAEQGLDGVAQKHLGPEKAALARQGRELFFAPAHLARRQEMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDAGTGKVSEPAVFTTRDMLRIE
YHMAESARRLSNRSGYAVSRKHVAAAIERIESDDPKNLFRLDAEQVDAIRHVTGDSGTAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWSEARRKAFGRSVAEGRDGHLRGDELLVLAHTNADVHKLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLLSVLLDNGRNVVFSEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAAKEDFAARPEWGHKARVDHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGSVHRLWGVSLPKA
LEQAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLARSGSKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGFAIERERRVAYNEERSETMIGAISSEERYLMPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASYTGTEPRGLADNLAVAPDRLGRLRGSELIVEGRAARDERDVAMA
ALEELLPLVRGHATEFRRNAERFELREQTRRASMSLSVPALSEQAMARLMEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELIRLFEAVKRFAEEQH
LAERRDRSKIVAAASAVPGAAPGKQTVTVLPMLAAVTEFKTSVDDEARSRAIAAPLYRQQRAALAEAARM
IWRDPAATVDNIESLLEKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARL
RSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEVKNNGRTRNAAAGSLDPSIHREFD
AVSRALDERFGRSAIVRGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRSRTVNRGRG
IEL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELMHEELALPDQIPAWLKMAMDG
RSIAKASEALWNAVDAFEKRADAQLARELIIALPEELTRAENIALVREFVSGNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGPKKVAVLGEDGKPLRVVTPDRPNGRIVYKVWAGDKETMKAWKVAWAETANR
HLALAGHDIRLDGRSYAEQGLDGVAQKHLGPEKAALARQGRELFFAPAHLARRQEMADRLLAEPELLLKQ
LGNERSTFDEKDIAKALHRYVDDPVEFANIRARLMASDELVMLKPQEMDAGTGKVSEPAVFTTRDMLRIE
YHMAESARRLSNRSGYAVSRKHVAAAIERIESDDPKNLFRLDAEQVDAIRHVTGDSGTAAVVGLAGAGKS
TLLAAARLAWESEGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELTWANGRDTLHRGDVLVIDEAGMVS
SQQMARVLKIAEEAEIKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRQEWARDASRLFARGE
VEKGLDAYAQQGHLVEAGTREEAINRIVADWSEARRKAFGRSVAEGRDGHLRGDELLVLAHTNADVHKLN
EALREVMKAEQALSDSRTFQTARGAREFAVGDRIIFLENARFLEPRARHSGPQYVKNGMLGTVVATGDKR
GDPLLSVLLDNGRNVVFSEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDRHLTYVSMTRHRDRADL
YAAKEDFAARPEWGHKARVDHAAGVTGELVEAGEAKFRPDDEDADDSPYADIRTDDGSVHRLWGVSLPKA
LEQAGISEGDTITLRKDGVERVKVQIAVVDEATGQKRYEEREVGRNVWTAKQIETAEARQERIERESHRP
DVFSRLVERLARSGSKTTTLDFESEAGYRAHADDFARRRGLDHLSLAAAEMEEGVSRRLAWIAEKRGQIA
KLWERASVALGFAIERERRVAYNEERSETMIGAISSEERYLMPPATDFRRSLDEDARLAQLSSPAWKERE
AILRPLLETIYRDPDAALVRLNALASYTGTEPRGLADNLAVAPDRLGRLRGSELIVEGRAARDERDVAMA
ALEELLPLVRGHATEFRRNAERFELREQTRRASMSLSVPALSEQAMARLMEIEAVRTQGGDDAYKTAFAL
AAEDRSVVQEVKAVSEALTARFGWSAFTAKADAMAERNMVERMPEDLSAGRREELIRLFEAVKRFAEEQH
LAERRDRSKIVAAASAVPGAAPGKQTVTVLPMLAAVTEFKTSVDDEARSRAIAAPLYRQQRAALAEAARM
IWRDPAATVDNIESLLEKGFAADRIAAAVTNDPAAYGALRGSDRLMDRMLASGRERKDAVQAVPEAAARL
RSLGSAYVNVLDAERQAIAEERRRMAVAIPGLSKAAEGVLMRLTTEVKNNGRTRNAAAGSLDPSIHREFD
AVSRALDERFGRSAIVRGEKDLLNRMPPAQRAAFAAMQEKLKVLQQTVRWESSEKIVSELRSRTVNRGRG
IEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1444 | GenBank | WP_046977229 |
| Name | traG_RPHASCH2410_RS29720_pRphCh2410d |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70437.30 Da Isoelectric Point: 9.2555
>WP_046977229.1 Ti-type conjugative transfer system protein TraG [Rhizobium phaseoli]
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSASYAMREFARLGPSVQTGVMTWDRALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
MIARGKPHPSLLFVFAPVVVTAIAVYIAGWRWPGLAAGMPGRMEYWFLRAAPVPALLFGPLAGLLTVWAL
PLHRRRPVAMTSLLCYLGVSASYAMREFARLGPSVQTGVMTWDRALSYLDMVAVLGAVAGFLAVAVSARI
SVVVPDEINRARRGTFGDADWLSMAAAAKLFPPDGEIVVGERYRVDKEIVHQLPFDANDRGTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTHALGREVMVLDPT
NPIMGFNVLDGIETSRQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAVKALDTARTVSAQCGEMTVEVKG
SSRNLGWNSRNGGTRKSESVSFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDGVAK
ANRFVKPVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 983749..993305
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RPHASCH2410_RS29755 (RPHASCH2410_PD04885) | 979319..979501 | + | 183 | WP_016735269 | hypothetical protein | - |
| RPHASCH2410_RS29760 (RPHASCH2410_PD04890) | 979634..980674 | - | 1041 | WP_016735270 | toprim domain-containing protein | - |
| RPHASCH2410_RS29765 (RPHASCH2410_PD04895) | 981037..982227 | + | 1191 | WP_016735272 | DUF1173 domain-containing protein | - |
| RPHASCH2410_RS29770 (RPHASCH2410_PD04900) | 982670..983263 | + | 594 | WP_037146723 | DUF433 domain-containing protein | - |
| RPHASCH2410_RS29775 (RPHASCH2410_PD04905) | 983278..983730 | + | 453 | WP_016735273 | PIN domain-containing protein | - |
| RPHASCH2410_RS29780 (RPHASCH2410_PD04910) | 983749..984744 | - | 996 | WP_037146721 | P-type DNA transfer ATPase VirB11 | virB11 |
| RPHASCH2410_RS29785 (RPHASCH2410_PD04915) | 984753..985928 | - | 1176 | WP_016735275 | type IV secretion system protein VirB10 | virB10 |
| RPHASCH2410_RS29790 (RPHASCH2410_PD04920) | 985937..986791 | - | 855 | WP_016735276 | P-type conjugative transfer protein VirB9 | virB9 |
| RPHASCH2410_RS29795 (RPHASCH2410_PD04925) | 986788..987459 | - | 672 | WP_037146717 | virB8 family protein | virB8 |
| RPHASCH2410_RS35430 (RPHASCH2410_PD04930) | 987453..987740 | - | 288 | WP_016737562 | hypothetical protein | - |
| RPHASCH2410_RS29805 (RPHASCH2410_PD04935) | 987781..988713 | - | 933 | WP_046977230 | type IV secretion system protein | virB6 |
| RPHASCH2410_RS29810 (RPHASCH2410_PD04940) | 988716..988949 | - | 234 | WP_016735278 | EexN family lipoprotein | - |
| RPHASCH2410_RS29815 (RPHASCH2410_PD04945) | 988946..989647 | - | 702 | WP_016735279 | P-type DNA transfer protein VirB5 | virB5 |
| RPHASCH2410_RS29820 (RPHASCH2410_PD04950) | 989644..992010 | - | 2367 | WP_046977231 | VirB4 family type IV secretion system protein | virb4 |
| RPHASCH2410_RS29825 (RPHASCH2410_PD04955) | 992003..992341 | - | 339 | WP_016735282 | type IV secretion system protein VirB3 | virB3 |
| RPHASCH2410_RS29830 (RPHASCH2410_PD04960) | 992347..992646 | - | 300 | WP_016735283 | TrbC/VirB2 family protein | virB2 |
| RPHASCH2410_RS29835 (RPHASCH2410_PD04965) | 992643..993305 | - | 663 | WP_016735284 | transglycosylase SLT domain-containing protein | virB1 |
| RPHASCH2410_RS29840 (RPHASCH2410_PD04970) | 993309..993845 | - | 537 | WP_016735285 | hypothetical protein | - |
| RPHASCH2410_RS29845 (RPHASCH2410_PD04975) | 993944..994315 | + | 372 | WP_016735286 | hypothetical protein | - |
| RPHASCH2410_RS29850 (RPHASCH2410_PD04985) | 994668..995858 | - | 1191 | WP_016735288 | hypothetical protein | - |
| RPHASCH2410_RS29855 (RPHASCH2410_PD04990) | 995932..996588 | - | 657 | WP_016735289 | hypothetical protein | - |
Host bacterium
| ID | 2932 | GenBank | NZ_AHJU02000028 |
| Plasmid name | pRphCh2410d | Incompatibility group | - |
| Plasmid size | 1099763 bp | Coordinate of oriT [Strand] | 972323..972351 [-] |
| Host baterium | Rhizobium phaseoli Ch24-10 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gmd |
| Metal resistance gene | hupE2 |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | AcrIIA8 |