Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102485 |
| Name | oriT_INSLA289|unnamed INSLA289_contig_133 |
| Organism | Escherichia coli strain INSLA289 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LHAT01000133 (1363..1435 [-], 73 nt) |
| oriT length | 73 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 73 nt
>oriT_INSLA289|unnamed INSLA289_contig_133
GTCGGGGCAAAGCCCTGACCAAGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
GTCGGGGCAAAGCCCTGACCAAGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file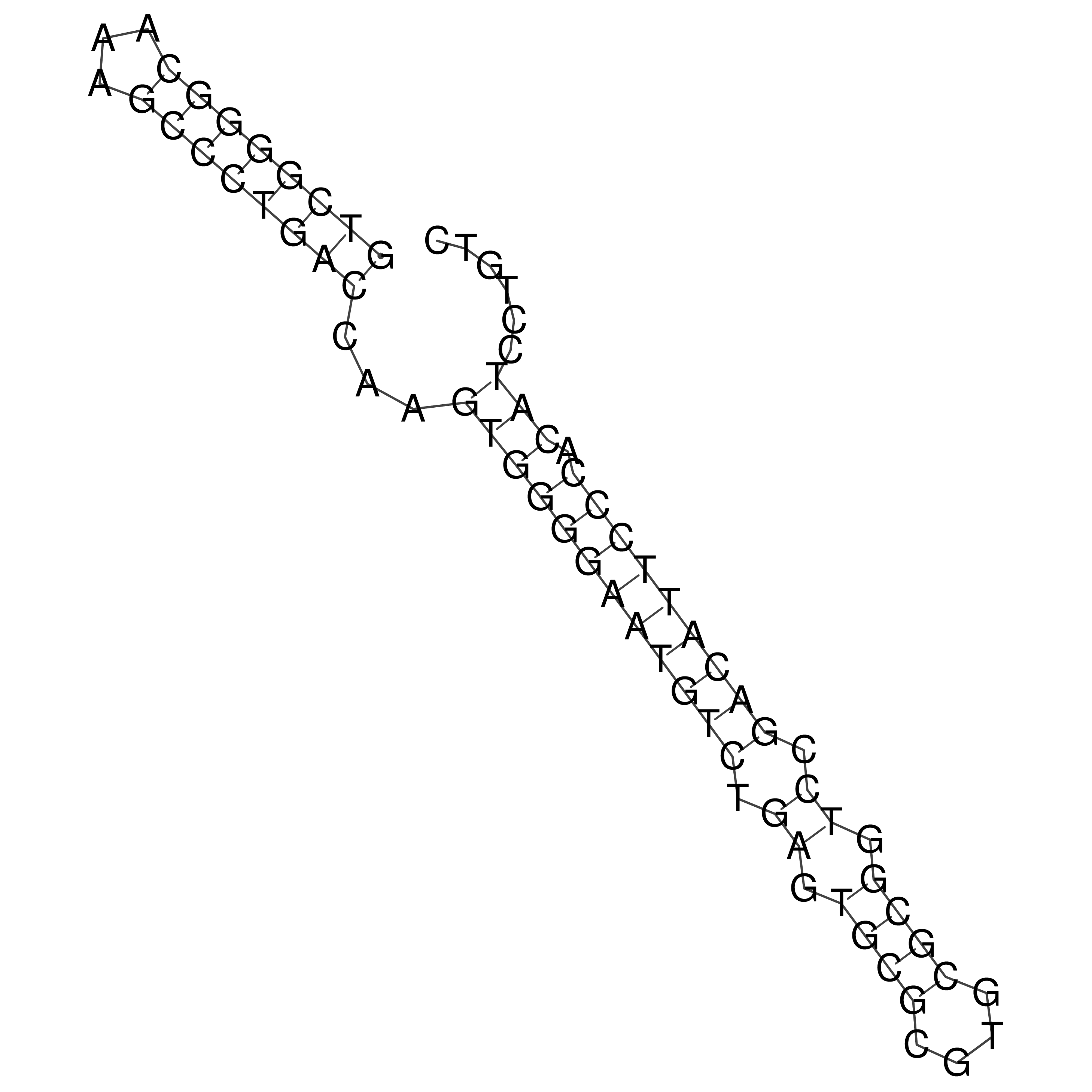
Relaxase
| ID | 1849 | GenBank | WP_021532735 |
| Name | Relaxase_AKG99_RS26180_INSLA289|unnamed INSLA289_contig_133 |
UniProt ID | _ |
| Length | 906 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 105183.62 Da Isoelectric Point: 8.3659
>WP_021532735.1 relaxase/mobilization nuclease domain-containing protein [Escherichia coli]
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGAPGRYEPEAINVDIRPEKVAETE
SLKQYACRHFAERLPKMARNGELKSCLDVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWHSVPKDIFTQVIPGERFRGHSIGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPELRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAHIRVQFRDPQLRKLHYHIAEVQRMQALIRLKENVREERLALMAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGAPGRYEPEAINVDIRPEKVAETE
SLKQYACRHFAERLPKMARNGELKSCLDVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWHSVPKDIFTQVIPGERFRGHSIGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPELRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAHIRVQFRDPQLRKLHYHIAEVQRMQALIRLKENVREERLALMAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 764 | GenBank | WP_001291058 |
| Name | WP_001291058_INSLA289|unnamed INSLA289_contig_133 |
UniProt ID | A0A7I0L241 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12730.52 Da Isoelectric Point: 9.9656
>WP_001291058.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0L241 |
T4CP
| ID | 1441 | GenBank | WP_021532734 |
| Name | trbC_AKG99_RS26185_INSLA289|unnamed INSLA289_contig_133 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87079.01 Da Isoelectric Point: 6.7760
>WP_021532734.1 F-type conjugative transfer protein TrbC [Escherichia coli]
MSQHHVNPDLIHRTAWGNPLWNALHNLNIIGLCLAGSIITALIWPLALPVCLLFTLVTSVIFTLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETSPARGIFYVGYRRINDIGRELWLSMDDLTRHII
FFSTTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEV
IQSGDKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QLLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFITQQSMILARDLGYRLE
GTDAQTLKVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMMAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYIAIEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
MSQHHVNPDLIHRTAWGNPLWNALHNLNIIGLCLAGSIITALIWPLALPVCLLFTLVTSVIFTLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETSPARGIFYVGYRRINDIGRELWLSMDDLTRHII
FFSTTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEV
IQSGDKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QLLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFITQQSMILARDLGYRLE
GTDAQTLKVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMMAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYIAIEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 7022..28942
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| AKG99_RS26185 (AKG99_26285) | 4741..7041 | - | 2301 | WP_021532734 | F-type conjugative transfer protein TrbC | - |
| AKG99_RS26190 (AKG99_26290) | 7022..8146 | - | 1125 | WP_021532733 | DsbC family protein | trbB |
| AKG99_RS26195 (AKG99_26295) | 8143..9405 | - | 1263 | WP_045145416 | IncI1-type conjugal transfer protein TrbA | trbA |
| AKG99_RS26200 (AKG99_26300) | 10058..10537 | - | 480 | WP_021532731 | thermonuclease family protein | - |
| AKG99_RS26205 (AKG99_26305) | 10702..11325 | + | 624 | WP_001155252 | helix-turn-helix domain-containing protein | - |
| AKG99_RS26210 (AKG99_26310) | 11523..11738 | - | 216 | WP_021532730 | hypothetical protein | - |
| AKG99_RS26215 (AKG99_26315) | 11742..12110 | - | 369 | WP_021532729 | hypothetical protein | - |
| AKG99_RS26220 (AKG99_26320) | 12122..12313 | - | 192 | WP_024187406 | hypothetical protein | - |
| AKG99_RS26225 (AKG99_26325) | 12409..12651 | - | 243 | WP_000650933 | hypothetical protein | - |
| AKG99_RS30800 | 12980..13132 | + | 153 | WP_021532728 | Hok/Gef family protein | - |
| AKG99_RS26230 (AKG99_26330) | 13185..13367 | - | 183 | WP_021532727 | hypothetical protein | - |
| AKG99_RS26235 (AKG99_26335) | 13701..13853 | + | 153 | WP_001367784 | Hok/Gef family protein | - |
| AKG99_RS33620 | 14168..14299 | - | 132 | Protein_16 | ethanolamine utilization protein EutE | - |
| AKG99_RS26245 (AKG99_26345) | 14394..14603 | + | 210 | WP_000062946 | HEAT repeat domain-containing protein | - |
| AKG99_RS26250 (AKG99_26350) | 14668..15318 | - | 651 | WP_021532726 | plasmid IncI1-type surface exclusion protein ExcA | - |
| AKG99_RS26255 (AKG99_26355) | 15407..17572 | - | 2166 | WP_021532725 | DotA/TraY family protein | traY |
| AKG99_RS26260 (AKG99_26360) | 17645..18214 | - | 570 | WP_021532724 | conjugal transfer protein TraX | - |
| AKG99_RS26265 (AKG99_26365) | 18211..19416 | - | 1206 | WP_052908626 | conjugal transfer protein TraW | traW |
| AKG99_RS26270 (AKG99_26370) | 19377..19994 | - | 618 | WP_052908627 | hypothetical protein | traV |
| AKG99_RS26275 (AKG99_26375) | 19994..23038 | - | 3045 | WP_021532721 | hypothetical protein | traU |
| AKG99_RS26280 (AKG99_26380) | 23214..23594 | - | 381 | WP_000690886 | hypothetical protein | - |
| AKG99_RS33625 | 23737..23952 | - | 216 | WP_072132148 | hypothetical protein | - |
| AKG99_RS26290 (AKG99_26390) | 24007..24825 | - | 819 | WP_072132149 | conjugal transfer protein TraT | traT |
| AKG99_RS33900 (AKG99_26395) | 24740..24991 | - | 252 | WP_021532719 | hypothetical protein | - |
| AKG99_RS26300 (AKG99_26400) | 25048..25446 | - | 399 | WP_021532718 | DUF6750 family protein | traR |
| AKG99_RS26305 (AKG99_26405) | 25493..26023 | - | 531 | WP_000987022 | conjugal transfer protein TraQ | traQ |
| AKG99_RS26310 (AKG99_26410) | 26020..26733 | - | 714 | WP_021532717 | conjugal transfer protein TraP | traP |
| AKG99_RS26315 (AKG99_26415) | 26730..28061 | - | 1332 | WP_021532716 | conjugal transfer protein TraO | traO |
| AKG99_RS26320 (AKG99_26420) | 28065..28942 | - | 878 | WP_052908628 | DotH/IcmK family type IV secretion protein | traN |
Host bacterium
| ID | 2929 | GenBank | NZ_LHAT01000133 |
| Plasmid name | INSLA289|unnamed INSLA289_contig_133 | Incompatibility group | - |
| Plasmid size | 28942 bp | Coordinate of oriT [Strand] | 1363..1435 [-] |
| Host baterium | Escherichia coli strain INSLA289 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |