Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102465 |
| Name | oriT_S8_VPH_Chula|unnamed2 |
| Organism | Salmonella enterica strain S8_VPH_Chula |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JARVXA010000004 (31201..31301 [+], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 80..85, 91..96 (AAAAAA..TTTTTT) 20..26, 38..44 (TAAATCA..TGATTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_S8_VPH_Chula|unnamed2
TATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
TATTTATTTTTTTATTTTTTAAATCAGTGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file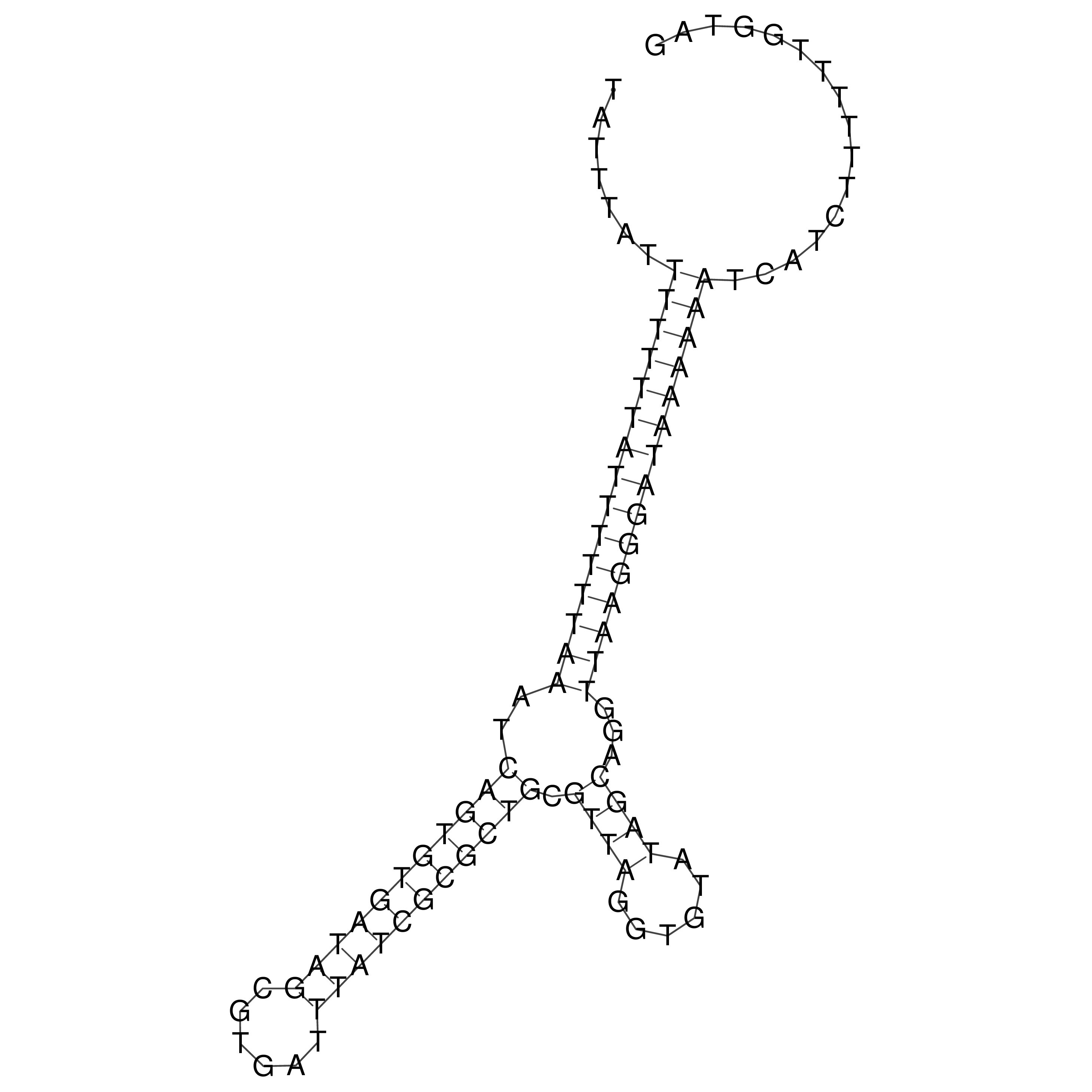
Relaxase
| ID | 1839 | GenBank | WP_011407157 |
| Name | mobF_QAT84_RS25525_S8_VPH_Chula|unnamed2 |
UniProt ID | A0A740S2B6 |
| Length | 1078 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1078 a.a. Molecular weight: 120179.41 Da Isoelectric Point: 6.5766
>WP_011407157.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLSGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLSGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A740S2B6 |
Auxiliary protein
| ID | 757 | GenBank | WP_000706094 |
| Name | WP_000706094_S8_VPH_Chula|unnamed2 |
UniProt ID | B6UZ32 |
| Length | 96 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 96 a.a. Molecular weight: 10829.54 Da Isoelectric Point: 8.5040
>WP_000706094.1 MULTISPECIES: hypothetical protein [Enterobacterales]
MKIVADRLSDVNRKLDYLFDRASDADFGPLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIA
EPGKTKAAKAEVERNGYKVWEPKKER
MKIVADRLSDVNRKLDYLFDRASDADFGPLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIA
EPGKTKAAKAEVERNGYKVWEPKKER
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B6UZ32 |
T4CP
| ID | 1428 | GenBank | WP_000342688 |
| Name | t4cp2_QAT84_RS25530_S8_VPH_Chula|unnamed2 |
UniProt ID | _ |
| Length | 509 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 509 a.a. Molecular weight: 57762.87 Da Isoelectric Point: 9.6551
>WP_000342688.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 14211..30634
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QAT84_RS25415 (QAT84_25415) | 10080..10637 | + | 558 | WP_001235713 | recombinase family protein | - |
| QAT84_RS25420 (QAT84_25420) | 10820..11680 | + | 861 | WP_000027057 | broad-spectrum class A beta-lactamase TEM-1 | - |
| QAT84_RS25425 (QAT84_25425) | 11881..12078 | - | 198 | WP_000844102 | hypothetical protein | - |
| QAT84_RS25430 (QAT84_25430) | 12083..12592 | - | 510 | WP_000893479 | restriction endonuclease | - |
| QAT84_RS25435 (QAT84_25435) | 12780..13091 | - | 312 | WP_001452736 | hypothetical protein | - |
| QAT84_RS25440 (QAT84_25440) | 13127..13441 | - | 315 | WP_000734776 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| QAT84_RS25445 (QAT84_25445) | 13404..13781 | - | 378 | WP_000504251 | hypothetical protein | - |
| QAT84_RS25450 (QAT84_25450) | 13797..14168 | - | 372 | WP_011867773 | H-NS family nucleoid-associated regulatory protein | - |
| QAT84_RS25455 (QAT84_25455) | 14211..14945 | + | 735 | WP_000033783 | lytic transglycosylase domain-containing protein | virB1 |
| QAT84_RS25460 (QAT84_25460) | 14954..15235 | + | 282 | WP_000440698 | transcriptional repressor KorA | - |
| QAT84_RS25465 (QAT84_25465) | 15245..15538 | + | 294 | WP_000209137 | TrbC/VirB2 family protein | virB2 |
| QAT84_RS25470 (QAT84_25470) | 15588..15905 | + | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| QAT84_RS25475 (QAT84_25475) | 15905..18505 | + | 2601 | WP_001200711 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| QAT84_RS25480 (QAT84_25480) | 18523..19236 | + | 714 | WP_000749362 | type IV secretion system protein | virB5 |
| QAT84_RS25485 (QAT84_25485) | 19244..19471 | + | 228 | WP_000734973 | IncN-type entry exclusion lipoprotein EexN | - |
| QAT84_RS25490 (QAT84_25490) | 19487..20527 | + | 1041 | WP_000886022 | type IV secretion system protein | virB6 |
| QAT84_RS25780 | 20628..20756 | + | 129 | WP_071882546 | conjugal transfer protein TraN | - |
| QAT84_RS25495 (QAT84_25495) | 20746..21444 | + | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| QAT84_RS25500 (QAT84_25500) | 21455..22339 | + | 885 | WP_000735067 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| QAT84_RS25505 (QAT84_25505) | 22339..23499 | + | 1161 | WP_000101711 | type IV secretion system protein VirB10 | virB10 |
| QAT84_RS25510 (QAT84_25510) | 23541..24536 | + | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| QAT84_RS25515 (QAT84_25515) | 24536..25069 | + | 534 | WP_000731968 | phospholipase D family protein | - |
| QAT84_RS25520 (QAT84_25520) | 25243..25869 | - | 627 | WP_000434930 | DUF6710 family protein | - |
| QAT84_RS25525 (QAT84_25525) | 25869..29105 | - | 3237 | WP_011407157 | MobF family relaxase | - |
| QAT84_RS25530 (QAT84_25530) | 29105..30634 | - | 1530 | WP_000342688 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| QAT84_RS25535 (QAT84_25535) | 30636..30926 | - | 291 | WP_000706094 | hypothetical protein | - |
| QAT84_RS25540 (QAT84_25540) | 31565..31984 | + | 420 | WP_000802909 | plasmid stabilization protein StbA | - |
| QAT84_RS25545 (QAT84_25545) | 31993..32709 | + | 717 | WP_000861577 | StbB family protein | - |
| QAT84_RS25550 (QAT84_25550) | 32711..33079 | + | 369 | WP_000414913 | plasmid stabilization protein StbC | - |
| QAT84_RS25555 (QAT84_25555) | 33261..33605 | + | 345 | WP_015059326 | hypothetical protein | - |
| QAT84_RS25560 (QAT84_25560) | 33716..34036 | - | 321 | WP_000211833 | protein CcgAII | - |
| QAT84_RS25565 (QAT84_25565) | 34091..34270 | - | 180 | WP_000214483 | protein CcgAI | - |
| QAT84_RS25570 (QAT84_25570) | 35155..35394 | - | 240 | WP_015059329 | hypothetical protein | - |
Host bacterium
| ID | 2909 | GenBank | NZ_JARVXA010000004 |
| Plasmid name | S8_VPH_Chula|unnamed2 | Incompatibility group | IncN |
| Plasmid size | 40254 bp | Coordinate of oriT [Strand] | 31201..31301 [+] |
| Host baterium | Salmonella enterica strain S8_VPH_Chula |
Cargo genes
| Drug resistance gene | qepA1, blaTEM-1B |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |