Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102464 |
| Name | oriT_S8_VPH_Chula|unnamed1 |
| Organism | Salmonella enterica strain S8_VPH_Chula |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JARVXA010000002 (91147..91270 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file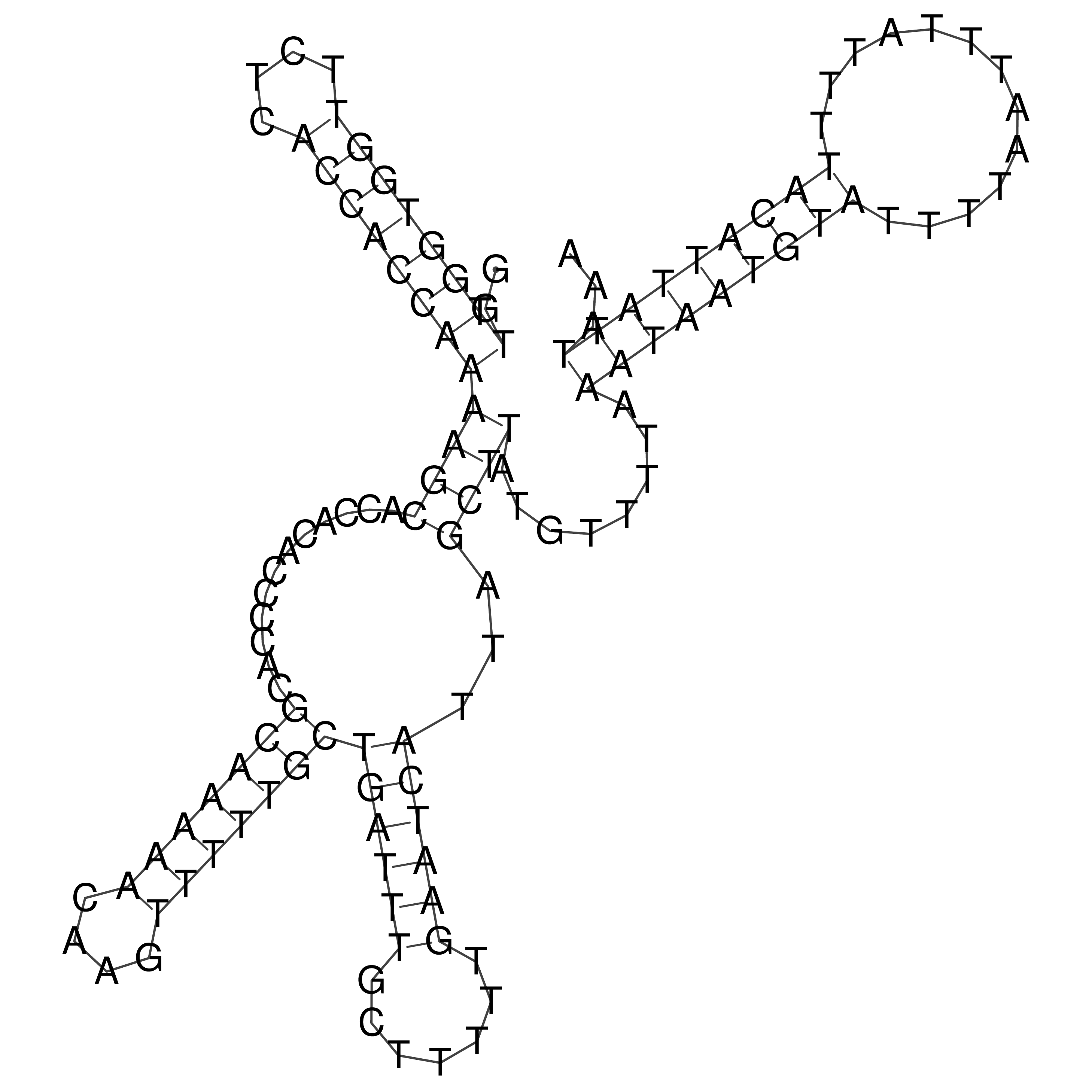
Relaxase
| ID | 1838 | GenBank | WP_279751861 |
| Name | mobF_QAT84_RS24630_S8_VPH_Chula|unnamed1 |
UniProt ID | _ |
| Length | 157 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 157 a.a. Molecular weight: 17124.20 Da Isoelectric Point: 6.4789
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQGHCCK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 755 | GenBank | WP_000124981 |
| Name | WP_000124981_S8_VPH_Chula|unnamed1 |
UniProt ID | A0A630FTW9 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14542.59 Da Isoelectric Point: 5.0278
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A630FTW9 |
| ID | 756 | GenBank | WP_001254386 |
| Name | WP_001254386_S8_VPH_Chula|unnamed1 |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
T4CP
| ID | 1426 | GenBank | WP_001496532 |
| Name | traD_QAT84_RS24635_S8_VPH_Chula|unnamed1 |
UniProt ID | _ |
| Length | 732 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 732 a.a. Molecular weight: 83213.84 Da Isoelectric Point: 5.0524
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTENPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQEN
HPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1427 | GenBank | WP_000069764 |
| Name | traC_QAT84_RS24715_S8_VPH_Chula|unnamed1 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99302.93 Da Isoelectric Point: 5.8486
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAA
ATQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLREKGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQY
TANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 63373..95102
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QAT84_RS24625 (QAT84_24625) | 62149..62853 | - | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| QAT84_RS24630 (QAT84_24630) | 62900..63373 | - | 474 | WP_279751861 | MobF family relaxase | - |
| QAT84_RS24635 (QAT84_24635) | 63373..65571 | - | 2199 | WP_001496532 | type IV conjugative transfer system coupling protein TraD | virb4 |
| QAT84_RS24640 (QAT84_24640) | 65824..66555 | - | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| QAT84_RS24645 (QAT84_24645) | 66569..67078 | - | 510 | WP_031943494 | conjugal transfer entry exclusion protein TraS | - |
| QAT84_RS24650 (QAT84_24650) | 67075..69900 | - | 2826 | WP_001007045 | conjugal transfer mating-pair stabilization protein TraG | traG |
| QAT84_RS24655 (QAT84_24655) | 69897..71273 | - | 1377 | WP_029400169 | conjugal transfer pilus assembly protein TraH | traH |
| QAT84_RS24660 (QAT84_24660) | 71270..71632 | - | 363 | WP_010379880 | P-type conjugative transfer protein TrbJ | - |
| QAT84_RS24665 (QAT84_24665) | 71562..72107 | - | 546 | WP_000059821 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| QAT84_RS24670 (QAT84_24670) | 72094..72378 | - | 285 | WP_000624109 | type-F conjugative transfer system pilin chaperone TraQ | - |
| QAT84_RS24675 (QAT84_24675) | 72497..72844 | - | 348 | WP_001287891 | conjugal transfer protein TrbA | - |
| QAT84_RS24680 (QAT84_24680) | 72860..73603 | - | 744 | WP_001430248 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| QAT84_RS24685 (QAT84_24685) | 73596..73853 | - | 258 | WP_000864352 | conjugal transfer protein TrbE | - |
| QAT84_RS24690 (QAT84_24690) | 73880..75688 | - | 1809 | WP_000821821 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| QAT84_RS24695 (QAT84_24695) | 75685..76323 | - | 639 | WP_000777692 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| QAT84_RS24700 (QAT84_24700) | 76332..77324 | - | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| QAT84_RS24705 (QAT84_24705) | 77321..77953 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| QAT84_RS24710 (QAT84_24710) | 77950..78336 | - | 387 | WP_000214092 | type-F conjugative transfer system protein TrbI | - |
| QAT84_RS24715 (QAT84_24715) | 78333..80963 | - | 2631 | WP_000069764 | type IV secretion system protein TraC | virb4 |
| QAT84_RS24720 (QAT84_24720) | 81089..81451 | - | 363 | WP_000836675 | hypothetical protein | - |
| QAT84_RS24725 (QAT84_24725) | 81479..81693 | - | 215 | Protein_101 | hypothetical protein | - |
| QAT84_RS24730 (QAT84_24730) | 81773..82249 | - | 477 | WP_015059129 | hypothetical protein | - |
| QAT84_RS24735 (QAT84_24735) | 82242..82463 | - | 222 | WP_001278695 | conjugal transfer protein TraR | - |
| QAT84_RS24740 (QAT84_24740) | 82598..83113 | - | 516 | WP_015059130 | type IV conjugative transfer system lipoprotein TraV | traV |
| QAT84_RS24745 (QAT84_24745) | 83110..83361 | - | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| QAT84_RS24750 (QAT84_24750) | 83354..83674 | - | 321 | WP_001057287 | conjugal transfer protein TrbD | virb4 |
| QAT84_RS24755 (QAT84_24755) | 83661..84251 | - | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| QAT84_RS24760 (QAT84_24760) | 84241..85341 | - | 1101 | Protein_108 | F-type conjugal transfer pilus assembly protein TraB | - |
| QAT84_RS24765 (QAT84_24765) | 85394..86098 | - | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| QAT84_RS24770 (QAT84_24770) | 86164..86487 | + | 324 | Protein_110 | hypothetical protein | - |
| QAT84_RS24775 (QAT84_24775) | 86542..86976 | + | 435 | WP_000845953 | conjugation system SOS inhibitor PsiB | - |
| QAT84_RS24780 (QAT84_24780) | 86973..87692 | + | 720 | WP_013362833 | plasmid SOS inhibition protein A | - |
| QAT84_RS24785 (QAT84_24785) | 87914..88063 | + | 150 | Protein_113 | plasmid maintenance protein Mok | - |
| QAT84_RS24790 (QAT84_24790) | 88005..88130 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| QAT84_RS24795 (QAT84_24795) | 88350..88580 | + | 231 | WP_001426396 | hypothetical protein | - |
| QAT84_RS24800 (QAT84_24800) | 88578..88751 | - | 174 | Protein_116 | hypothetical protein | - |
| QAT84_RS24805 (QAT84_24805) | 88821..89027 | + | 207 | WP_000547968 | hypothetical protein | - |
| QAT84_RS24810 (QAT84_24810) | 89052..89339 | + | 288 | WP_000107544 | hypothetical protein | - |
| QAT84_RS24815 (QAT84_24815) | 89457..90278 | + | 822 | WP_001234469 | DUF932 domain-containing protein | - |
| QAT84_RS24820 (QAT84_24820) | 90575..91222 | - | 648 | WP_031943493 | transglycosylase SLT domain-containing protein | virB1 |
| QAT84_RS24825 (QAT84_24825) | 91499..91882 | + | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| QAT84_RS24830 (QAT84_24830) | 92073..92759 | + | 687 | WP_000332484 | PAS domain-containing protein | - |
| QAT84_RS24835 (QAT84_24835) | 92853..93080 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| QAT84_RS24840 (QAT84_24840) | 93114..93473 | + | 360 | WP_000340272 | type IV conjugative transfer system pilin TraA | - |
| QAT84_RS24845 (QAT84_24845) | 93488..93799 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| QAT84_RS24850 (QAT84_24850) | 93821..94387 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| QAT84_RS24855 (QAT84_24855) | 94374..95102 | + | 729 | WP_001230813 | type-F conjugative transfer system secretin TraK | traK |
| QAT84_RS24860 (QAT84_24860) | 95102..95437 | + | 336 | Protein_128 | conjugal transfer protein TraB | - |
| QAT84_RS24865 (QAT84_24865) | 95500..96204 | + | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| QAT84_RS24870 (QAT84_24870) | 96255..96320 | - | 66 | Protein_130 | hypothetical protein | - |
| QAT84_RS24875 (QAT84_24875) | 96463..97323 | + | 861 | WP_000557454 | aminoglycoside N-acetyltransferase AAC(3)-IId | - |
| QAT84_RS24880 (QAT84_24880) | 97336..97878 | + | 543 | WP_000587837 | AAA family ATPase | - |
| QAT84_RS24885 (QAT84_24885) | 98021..98725 | - | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| QAT84_RS24890 (QAT84_24890) | 98786..99256 | + | 471 | Protein_134 | recombinase family protein | - |
Host bacterium
| ID | 2908 | GenBank | NZ_JARVXA010000002 |
| Plasmid name | S8_VPH_Chula|unnamed1 | Incompatibility group | IncA/C2 |
| Plasmid size | 142378 bp | Coordinate of oriT [Strand] | 91147..91270 [+] |
| Host baterium | Salmonella enterica strain S8_VPH_Chula |
Cargo genes
| Drug resistance gene | floR, tet(A), aph(6)-Id, aph(3'')-Ib, sul2, catA2, blaCTX-M-55, qnrS1, aac(3)-IId, blaTEM-1B |
| Virulence gene | - |
| Metal resistance gene | merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |