Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102420 |
| Name | oriT_SMG004F8|unnamed1 |
| Organism | Escherichia coli strain SMG004F8 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAKVSX010000022 (48287..48375 [-], 89 nt) |
| oriT length | 89 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 89 nt
>oriT_SMG004F8|unnamed1
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTC
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGTAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTACACATCCTGTCCCGTTTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file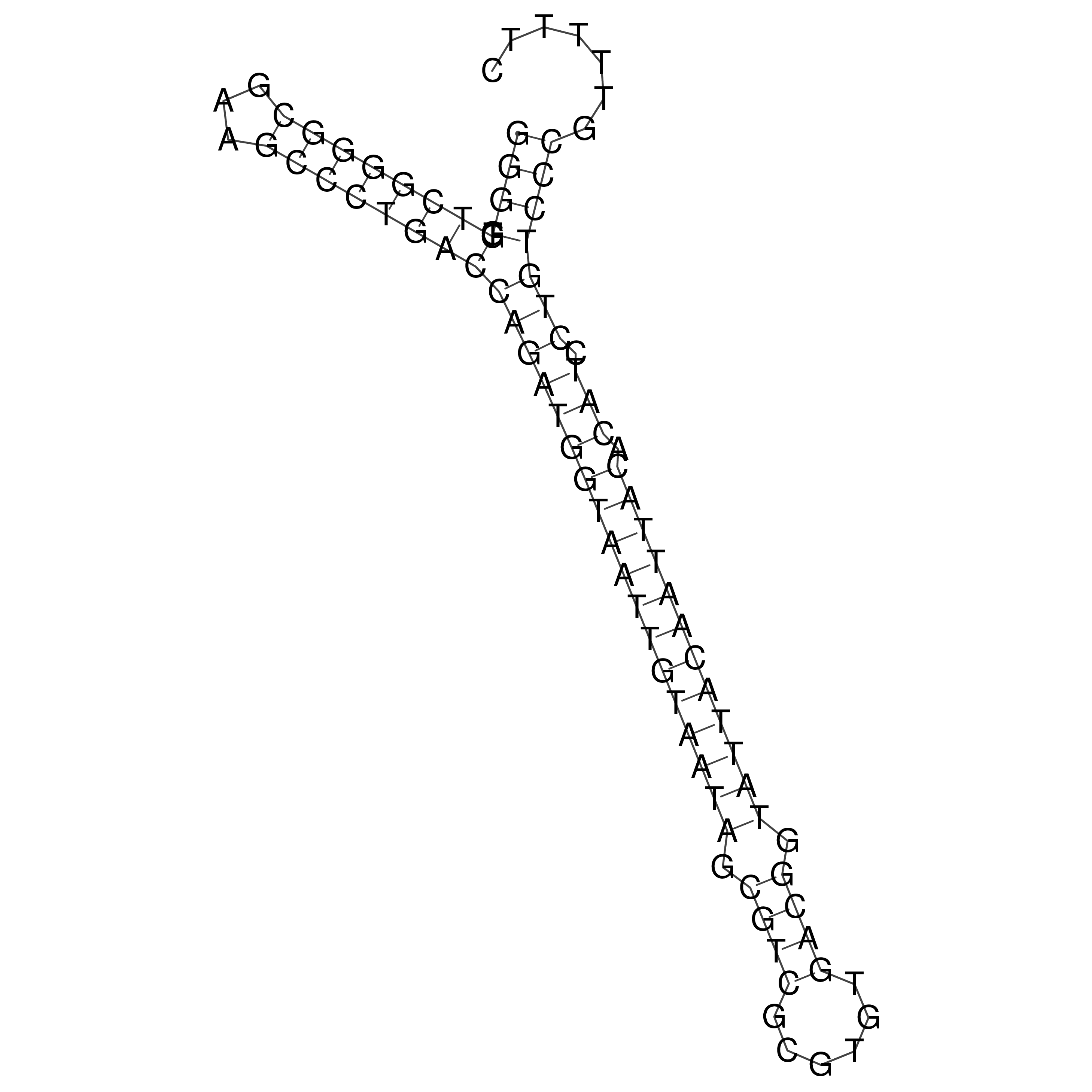
Relaxase
| ID | 1812 | GenBank | WP_024236743 |
| Name | nikB_ML391_RS19715_SMG004F8|unnamed1 |
UniProt ID | _ |
| Length | 898 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 898 a.a. Molecular weight: 103815.28 Da Isoelectric Point: 7.5153
>WP_024236743.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSQAEQPHRSRFSRLVDYATRLRNESFVALVDV
MKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVRH
TLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCWV
HAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQKDGKF
LVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETES
LQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLTL
DNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGYS
ISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEPQ
AGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRKP
DLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYPP
SYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGSV
RFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAWH
NVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELNLSSSQAEQPHRSRFSRLVDYATRLRNESFVALVDV
MKDGCEWVNFYGVTCFHNCTSLETAAADMEYIAQQAHYAKDNTDPVFHYILSWQAHESPRPEQIYDSVRH
TLKSLGLGEHQYVSAVHTDTDNLHVHVAVNRVHPVTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCWV
HAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQKDGKF
LVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETES
LQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLTL
DNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGYS
ISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEPQ
AGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRKP
DLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYPP
SYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGSV
RFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAWH
NVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 732 | GenBank | WP_001283947 |
| Name | WP_001283947_SMG004F8|unnamed1 |
UniProt ID | A0A142CMC2 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12613.57 Da Isoelectric Point: 10.7463
>WP_001283947.1 MULTISPECIES: IncI1-type relaxosome accessory protein NikA [Enterobacteriaceae]
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CMC2 |
T4CP
| ID | 1397 | GenBank | WP_011264051 |
| Name | trbC_ML391_RS19720_SMG004F8|unnamed1 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86954.10 Da Isoelectric Point: 6.7713
>WP_011264051.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKNNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKNNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 53750..83183
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ML391_RS19720 (ML391_19720) | 51466..53757 | - | 2292 | WP_011264051 | F-type conjugative transfer protein TrbC | - |
| ML391_RS19725 (ML391_19725) | 53750..54820 | - | 1071 | WP_024236742 | IncI1-type conjugal transfer protein TrbB | trbB |
| ML391_RS19730 (ML391_19730) | 54839..56047 | - | 1209 | WP_000121273 | IncI1-type conjugal transfer protein TrbA | trbA |
| ML391_RS19735 (ML391_19735) | 56339..56491 | + | 153 | WP_001387489 | Hok/Gef family protein | - |
| ML391_RS19740 (ML391_19740) | 56563..56814 | - | 252 | WP_001291964 | hypothetical protein | - |
| ML391_RS27690 | 57315..57410 | + | 96 | WP_001303310 | DinQ-like type I toxin DqlB | - |
| ML391_RS19750 (ML391_19750) | 57475..57651 | - | 177 | WP_001054897 | hypothetical protein | - |
| ML391_RS19755 (ML391_19755) | 58043..58252 | + | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| ML391_RS19760 (ML391_19760) | 58324..58974 | - | 651 | WP_021520375 | plasmid IncI1-type surface exclusion protein ExcA | - |
| ML391_RS19765 (ML391_19765) | 59048..61216 | - | 2169 | WP_000698357 | DotA/TraY family protein | traY |
| ML391_RS19770 (ML391_19770) | 61313..61897 | - | 585 | WP_001037987 | IncI1-type conjugal transfer protein TraX | - |
| ML391_RS19775 (ML391_19775) | 61926..63128 | - | 1203 | WP_001189160 | IncI1-type conjugal transfer protein TraW | traW |
| ML391_RS19780 (ML391_19780) | 63095..63709 | - | 615 | WP_000337398 | IncI1-type conjugal transfer protein TraV | traV |
| ML391_RS19785 (ML391_19785) | 63709..66753 | - | 3045 | WP_001024783 | IncI1-type conjugal transfer protein TraU | traU |
| ML391_RS19790 (ML391_19790) | 66843..67643 | - | 801 | WP_001164788 | IncI1-type conjugal transfer protein TraT | traT |
| ML391_RS19795 (ML391_19795) | 67627..67815 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| ML391_RS19800 (ML391_19800) | 67879..68283 | - | 405 | WP_000086959 | IncI1-type conjugal transfer protein TraR | traR |
| ML391_RS19805 (ML391_19805) | 68334..68861 | - | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| ML391_RS19810 (ML391_19810) | 68861..69565 | - | 705 | WP_000801919 | IncI1-type conjugal transfer protein TraP | traP |
| ML391_RS19815 (ML391_19815) | 69565..70854 | - | 1290 | WP_001272000 | conjugal transfer protein TraO | traO |
| ML391_RS19820 (ML391_19820) | 70857..71840 | - | 984 | WP_001191879 | IncI1-type conjugal transfer protein TraN | traN |
| ML391_RS19825 (ML391_19825) | 71851..72543 | - | 693 | WP_000138550 | DotI/IcmL family type IV secretion protein | traM |
| ML391_RS19830 (ML391_19830) | 72540..72887 | - | 348 | WP_001055900 | conjugal transfer protein | traL |
| ML391_RS19835 (ML391_19835) | 72905..76672 | - | 3768 | WP_021520378 | LPD7 domain-containing protein | - |
| ML391_RS19840 (ML391_19840) | 76762..77313 | - | 552 | WP_000014583 | phospholipase D family protein | - |
| ML391_RS19845 (ML391_19845) | 77328..77618 | - | 291 | WP_001299214 | hypothetical protein | traK |
| ML391_RS19850 (ML391_19850) | 77615..78763 | - | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| ML391_RS19855 (ML391_19855) | 78760..79578 | - | 819 | WP_000646098 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| ML391_RS19860 (ML391_19860) | 79575..80033 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| ML391_RS19865 (ML391_19865) | 80428..81012 | - | 585 | WP_000977522 | histidine phosphatase family protein | - |
| ML391_RS19870 (ML391_19870) | 81072..82274 | - | 1203 | WP_000976351 | conjugal transfer protein TraF | - |
| ML391_RS19875 (ML391_19875) | 82359..83183 | - | 825 | WP_001238939 | conjugal transfer protein TraE | traE |
| ML391_RS19880 (ML391_19880) | 83334..84488 | - | 1155 | WP_001139957 | site-specific integrase | - |
| ML391_RS27800 | 84487..84665 | + | 179 | Protein_97 | shufflon protein D' | - |
Host bacterium
| ID | 2864 | GenBank | NZ_JAKVSX010000022 |
| Plasmid name | SMG004F8|unnamed1 | Incompatibility group | IncI1 |
| Plasmid size | 84665 bp | Coordinate of oriT [Strand] | 48287..48375 [-] |
| Host baterium | Escherichia coli strain SMG004F8 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |