Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102417 |
| Name | oriT_SMG002F9|unnamed2 |
| Organism | Escherichia coli strain SMG002F9 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAKVSS010000026 (6685..6808 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACTAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file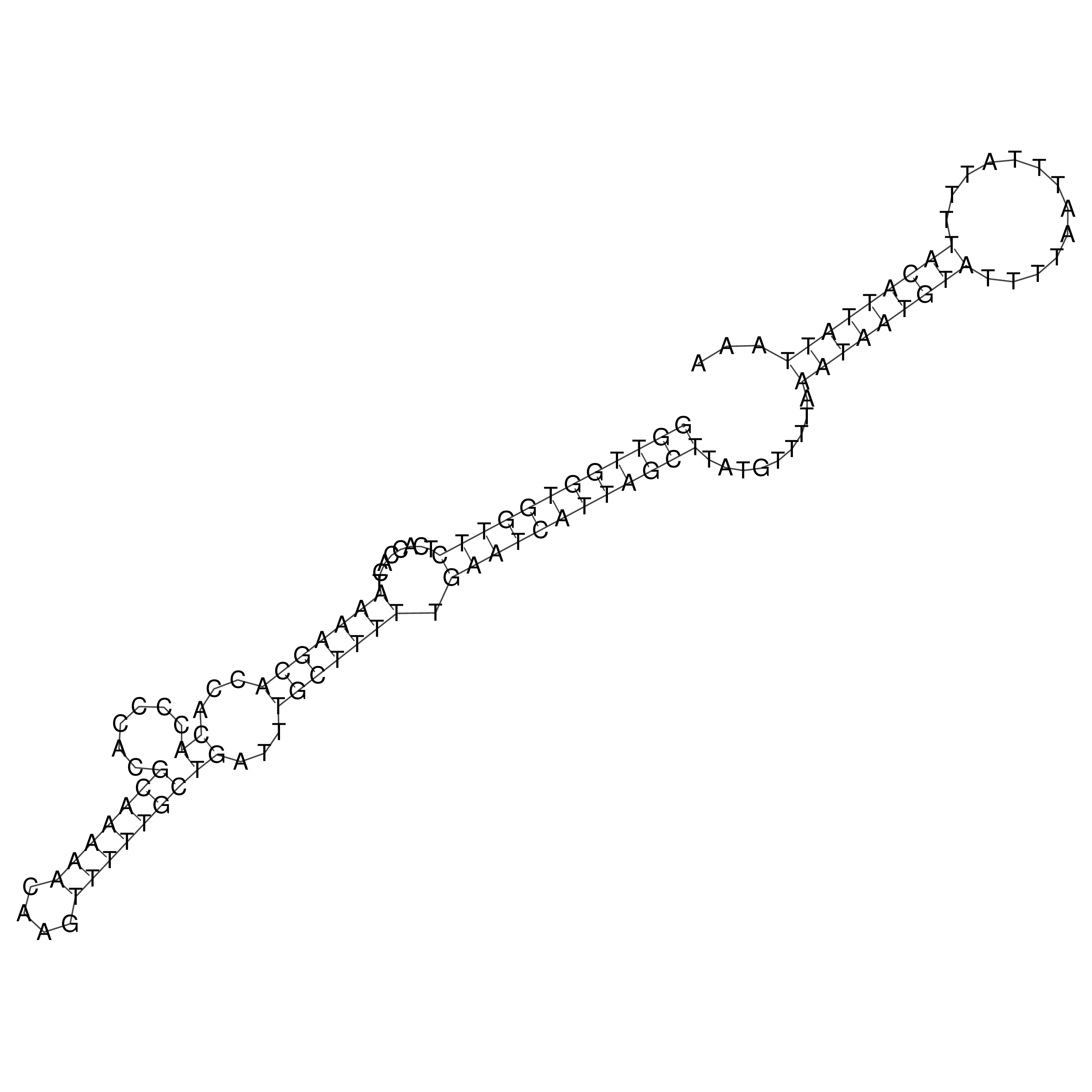
Relaxase
| ID | 1810 | GenBank | WP_053001887 |
| Name | traI_ML382_RS20300_SMG002F9|unnamed2 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192126.75 Da Isoelectric Point: 5.8440
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMVREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHQEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKISGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVVESKNLAFSMVDLLTEAKSFAAEGTSFADLGREINT
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETPDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWVPEHSVTEFSHSQEAKLAEA
QQEAIQKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALVLVDNVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGIRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DSKTEQAVREIAGQERDRSAISEREAALPESVLRESQREREAVREVARENLLQERLQQMEQDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 729 | GenBank | WP_000124981 |
| Name | WP_000124981_SMG002F9|unnamed2 |
UniProt ID | A0A630FTW9 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14542.59 Da Isoelectric Point: 5.0278
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYASMVDDIREKVSVEMDRFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A630FTW9 |
| ID | 730 | GenBank | WP_001254386 |
| Name | WP_001254386_SMG002F9|unnamed2 |
UniProt ID | A0A3Z6KJB5 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.11 Da Isoelectric Point: 10.1422
MRRRNARGGISRTVSVYLDEDTNNRLIKAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3Z6KJB5 |
T4CP
| ID | 1394 | GenBank | WP_001064268 |
| Name | traC_ML382_RS20205_SMG002F9|unnamed2 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99248.97 Da Isoelectric Point: 6.0801
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKNKARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1395 | GenBank | WP_001360340 |
| Name | traD_ML382_RS20295_SMG002F9|unnamed2 |
UniProt ID | _ |
| Length | 729 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 729 a.a. Molecular weight: 82860.46 Da Isoelectric Point: 5.0524
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTENPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPD
IQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6113..32776
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ML382_RS20085 (ML382_20080) | 2079..2513 | + | 435 | WP_096309212 | conjugation system SOS inhibitor PsiB | - |
| ML382_RS20090 (ML382_20085) | 2510..3272 | + | 763 | Protein_2 | plasmid SOS inhibition protein A | - |
| ML382_RS20095 (ML382_20090) | 3241..3429 | - | 189 | WP_001299721 | hypothetical protein | - |
| ML382_RS20100 (ML382_20095) | 3451..3600 | + | 150 | Protein_4 | plasmid maintenance protein Mok | - |
| ML382_RS20105 (ML382_20100) | 3542..3667 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| ML382_RS20110 (ML382_20105) | 3887..4117 | + | 231 | WP_072279680 | hypothetical protein | - |
| ML382_RS20115 (ML382_20110) | 4115..4288 | - | 174 | Protein_7 | hypothetical protein | - |
| ML382_RS20120 (ML382_20115) | 4358..4564 | + | 207 | WP_000547968 | hypothetical protein | - |
| ML382_RS20125 (ML382_20120) | 4589..4876 | + | 288 | WP_000107548 | hypothetical protein | - |
| ML382_RS20130 (ML382_20125) | 4995..5816 | + | 822 | WP_052953840 | DUF932 domain-containing protein | - |
| ML382_RS20135 (ML382_20130) | 6113..6760 | - | 648 | WP_001567347 | transglycosylase SLT domain-containing protein | virB1 |
| ML382_RS20140 (ML382_20135) | 7037..7420 | + | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| ML382_RS20145 (ML382_20140) | 7611..8297 | + | 687 | WP_000332484 | PAS domain-containing protein | - |
| ML382_RS20150 (ML382_20145) | 8391..8618 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| ML382_RS20155 (ML382_20150) | 8652..9014 | + | 363 | WP_033566647 | type IV conjugative transfer system pilin TraA | - |
| ML382_RS20160 (ML382_20155) | 9019..9330 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| ML382_RS20165 (ML382_20160) | 9352..9918 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| ML382_RS20170 (ML382_20165) | 9905..10633 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| ML382_RS20175 (ML382_20170) | 10633..12060 | + | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| ML382_RS20180 (ML382_20175) | 12050..12622 | + | 573 | WP_000002780 | conjugal transfer pilus-stabilizing protein TraP | - |
| ML382_RS20185 (ML382_20180) | 12619..12936 | + | 318 | WP_001057292 | conjugal transfer protein TrbD | virb4 |
| ML382_RS20190 (ML382_20185) | 12933..13184 | + | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| ML382_RS20195 (ML382_20190) | 13181..13696 | + | 516 | WP_032178525 | type IV conjugative transfer system lipoprotein TraV | traV |
| ML382_RS20200 (ML382_20195) | 13830..14051 | + | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| ML382_RS20205 (ML382_20200) | 14211..16838 | + | 2628 | WP_001064268 | type IV secretion system protein TraC | virb4 |
| ML382_RS20210 (ML382_20205) | 16835..17221 | + | 387 | WP_000214084 | type-F conjugative transfer system protein TrbI | - |
| ML382_RS20215 (ML382_20210) | 17218..17850 | + | 633 | WP_104159822 | type-F conjugative transfer system protein TraW | traW |
| ML382_RS20220 (ML382_20215) | 17847..18839 | + | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| ML382_RS20225 (ML382_20220) | 18865..19326 | + | 462 | WP_000277845 | hypothetical protein | - |
| ML382_RS20230 (ML382_20225) | 19356..19808 | + | 453 | WP_000069427 | hypothetical protein | - |
| ML382_RS20235 (ML382_20230) | 19836..20474 | + | 639 | WP_001104248 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| ML382_RS20240 (ML382_20235) | 20471..22279 | + | 1809 | WP_000821818 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| ML382_RS20245 (ML382_20240) | 22306..22563 | + | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| ML382_RS20250 (ML382_20245) | 22556..23299 | + | 744 | WP_021553228 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| ML382_RS20255 (ML382_20250) | 23315..23662 | + | 348 | WP_053001888 | conjugal transfer protein TrbA | - |
| ML382_RS20260 (ML382_20255) | 23781..24065 | + | 285 | WP_000624110 | type-F conjugative transfer system pilin chaperone TraQ | - |
| ML382_RS20265 (ML382_20260) | 24052..24597 | + | 546 | WP_000052618 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| ML382_RS20270 (ML382_20265) | 24527..24889 | + | 363 | WP_001443292 | P-type conjugative transfer protein TrbJ | - |
| ML382_RS20275 (ML382_20270) | 24889..26262 | + | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| ML382_RS20280 (ML382_20275) | 26259..29084 | + | 2826 | WP_001554363 | conjugal transfer mating-pair stabilization protein TraG | traG |
| ML382_RS20285 (ML382_20280) | 29081..29590 | + | 510 | WP_000628099 | conjugal transfer entry exclusion protein TraS | - |
| ML382_RS20290 (ML382_20285) | 29604..30335 | + | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| ML382_RS20295 (ML382_20290) | 30587..32776 | + | 2190 | WP_001360340 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2861 | GenBank | NZ_JAKVSS010000026 |
| Plasmid name | SMG002F9|unnamed2 | Incompatibility group | IncFII |
| Plasmid size | 61709 bp | Coordinate of oriT [Strand] | 6685..6808 [+] |
| Host baterium | Escherichia coli strain SMG002F9 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |