Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102350 |
| Name | oriT_pAR-0410 |
| Organism | Salmonella enterica subsp. enterica serovar Infantis strain AR-0410 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_WACL01000005 (283992..284080 [-], 89 nt) |
| oriT length | 89 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 89 nt
>oriT_pAR-0410
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file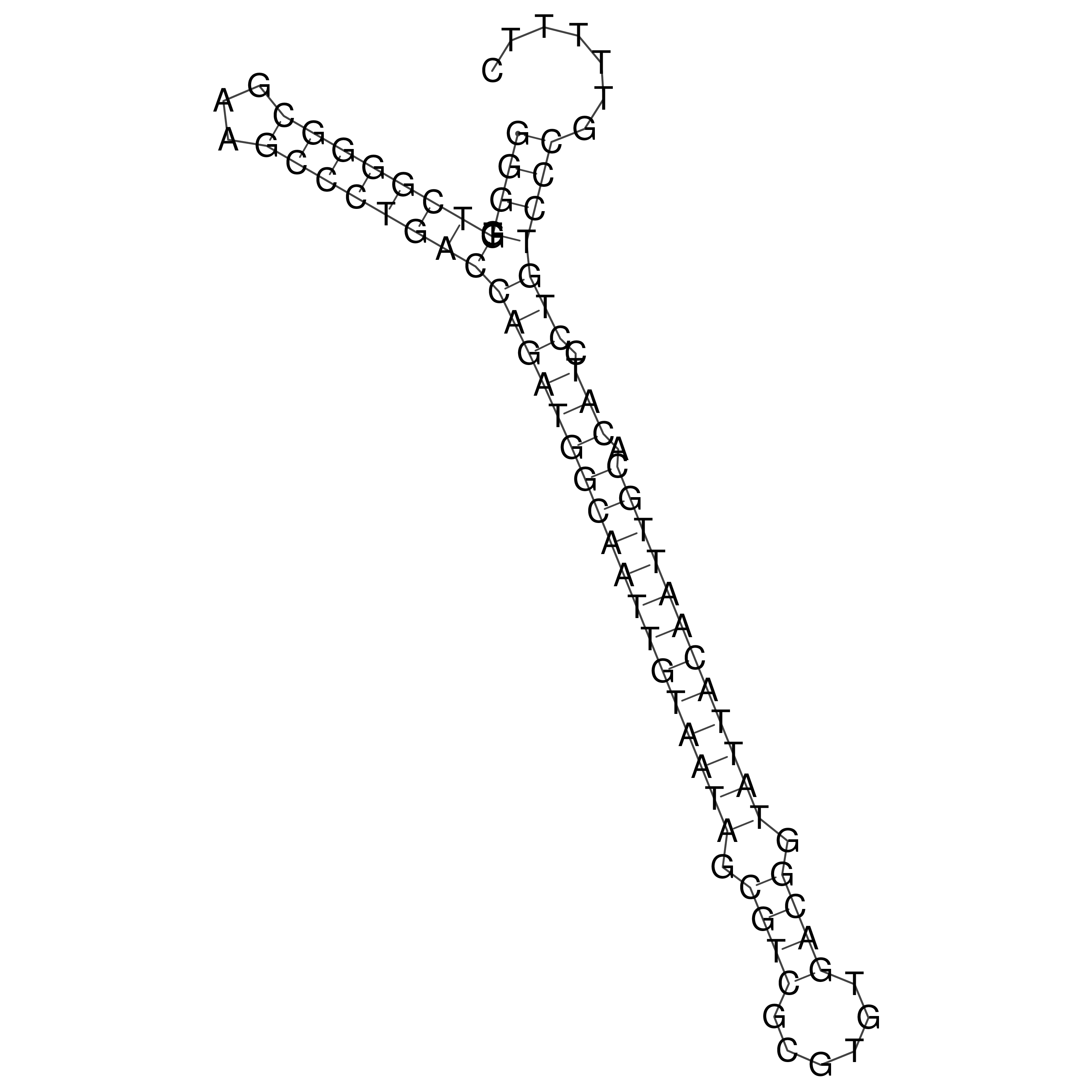
Relaxase
| ID | 1781 | GenBank | WP_006855641 |
| Name | nikB_F4V22_RS23490_pAR-0410 |
UniProt ID | A0A620WL05 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104006.42 Da Isoelectric Point: 7.3526
>WP_006855641.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLGERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWQPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A620WL05 |
Auxiliary protein
| ID | 701 | GenBank | WP_001283947 |
| Name | WP_001283947_pAR-0410 |
UniProt ID | A0A142CMC2 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12613.57 Da Isoelectric Point: 10.7463
>WP_001283947.1 MULTISPECIES: IncI1-type relaxosome accessory protein NikA [Enterobacteriaceae]
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDSAVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A142CMC2 |
T4CP
| ID | 1364 | GenBank | WP_001289275 |
| Name | trbC_F4V22_RS23495_pAR-0410 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86957.15 Da Isoelectric Point: 6.7713
>WP_001289275.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHLTGRPVNGFHHKKTNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLECADPSQDRMIKRSLFSFWPTLFQYEVILESPASGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHLTGRPVNGFHHKKTNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 289458..320322
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| F4V22_RS23495 | 287174..289465 | - | 2292 | WP_001289275 | F-type conjugative transfer protein TrbC | - |
| F4V22_RS23500 | 289458..290528 | - | 1071 | WP_000151583 | IncI1-type conjugal transfer protein TrbB | trbB |
| F4V22_RS23505 | 290547..291755 | - | 1209 | WP_001383960 | IncI1-type conjugal transfer protein TrbA | trbA |
| F4V22_RS23510 | 292062..292841 | - | 780 | WP_275450201 | protein FinQ | - |
| F4V22_RS23515 | 293468..293620 | + | 153 | WP_001387489 | Hok/Gef family protein | - |
| F4V22_RS23520 | 293692..293943 | - | 252 | WP_001291964 | hypothetical protein | - |
| F4V22_RS25550 | 294444..294539 | + | 96 | WP_000609148 | DinQ-like type I toxin DqlB | - |
| F4V22_RS25120 | 294604..294780 | - | 177 | WP_001054897 | hypothetical protein | - |
| F4V22_RS23530 | 295172..295381 | + | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| F4V22_RS23535 | 295453..296115 | - | 663 | WP_000644796 | plasmid IncI1-type surface exclusion protein ExcA | - |
| F4V22_RS23540 | 296186..298354 | - | 2169 | WP_058649949 | DotA/TraY family protein | traY |
| F4V22_RS23545 | 298451..299035 | - | 585 | WP_001037996 | IncI1-type conjugal transfer protein TraX | - |
| F4V22_RS23550 | 299064..300266 | - | 1203 | WP_001189156 | IncI1-type conjugal transfer protein TraW | traW |
| F4V22_RS23555 | 300233..300847 | - | 615 | WP_000337399 | IncI1-type conjugal transfer protein TraV | traV |
| F4V22_RS23560 | 300847..303891 | - | 3045 | WP_001024776 | IncI1-type conjugal transfer protein TraU | traU |
| F4V22_RS23565 | 303981..304781 | - | 801 | WP_001164792 | IncI1-type conjugal transfer protein TraT | traT |
| F4V22_RS23570 | 304765..304953 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| F4V22_RS23575 | 305017..305421 | - | 405 | WP_000086960 | IncI1-type conjugal transfer protein TraR | traR |
| F4V22_RS23580 | 305472..305999 | - | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| F4V22_RS23585 | 305999..306703 | - | 705 | WP_023155244 | IncI1-type conjugal transfer protein TraP | traP |
| F4V22_RS23590 | 306703..307992 | - | 1290 | WP_001271994 | conjugal transfer protein TraO | traO |
| F4V22_RS23595 | 307995..308978 | - | 984 | WP_021513963 | IncI1-type conjugal transfer protein TraN | traN |
| F4V22_RS23600 | 308989..309681 | - | 693 | WP_000138552 | DotI/IcmL family type IV secretion protein | traM |
| F4V22_RS23605 | 309678..310025 | - | 348 | WP_001055900 | conjugal transfer protein | traL |
| F4V22_RS23610 | 310043..313810 | - | 3768 | WP_058649948 | LPD7 domain-containing protein | - |
| F4V22_RS23615 | 313900..314451 | - | 552 | WP_000014584 | phospholipase D family protein | - |
| F4V22_RS23620 | 314466..314756 | - | 291 | WP_001299214 | hypothetical protein | traK |
| F4V22_RS23625 | 314753..315901 | - | 1149 | WP_001024976 | plasmid transfer ATPase TraJ | virB11 |
| F4V22_RS23630 | 315898..316716 | - | 819 | WP_000646097 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| F4V22_RS23635 | 316713..317171 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| F4V22_RS23640 | 317566..318150 | - | 585 | WP_000977520 | histidine phosphatase family protein | - |
| F4V22_RS23645 | 318210..319412 | - | 1203 | WP_000976353 | conjugal transfer protein TraF | - |
| F4V22_RS23650 | 319498..320322 | - | 825 | WP_058649947 | conjugal transfer protein TraE | traE |
| F4V22_RS23655 | 320473..321627 | - | 1155 | WP_001139955 | site-specific integrase | - |
Host bacterium
| ID | 2794 | GenBank | NZ_WACL01000005 |
| Plasmid name | pAR-0410 | Incompatibility group | IncFIB |
| Plasmid size | 321860 bp | Coordinate of oriT [Strand] | 283992..284080 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Infantis strain AR-0410 |
Cargo genes
| Drug resistance gene | tet(A), sul1, qacE, ant(3'')-Ia, aph(3')-Ia, dfrA14, aac(3)-IVa, aph(4)-Ia, floR, fosA3, blaCTX-M-65 |
| Virulence gene | ybtS, ybtX, ybtQ, ybtP, ybtA, irp2, ybtT, ybtE, fyuA, faeI, faeH, faeF, faeE, faeC |
| Metal resistance gene | merR, merT, merP, merA, merD, merE, arsA, arsH |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrVA2 |