Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102344 |
| Name | oriT_KZN_INI_KINF_24|unnamed45 |
| Organism | Klebsiella pneumoniae strain KZN_INI_KINF_24 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_VOOC01000099 (1994..2042 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_KZN_INI_KINF_24|unnamed45
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file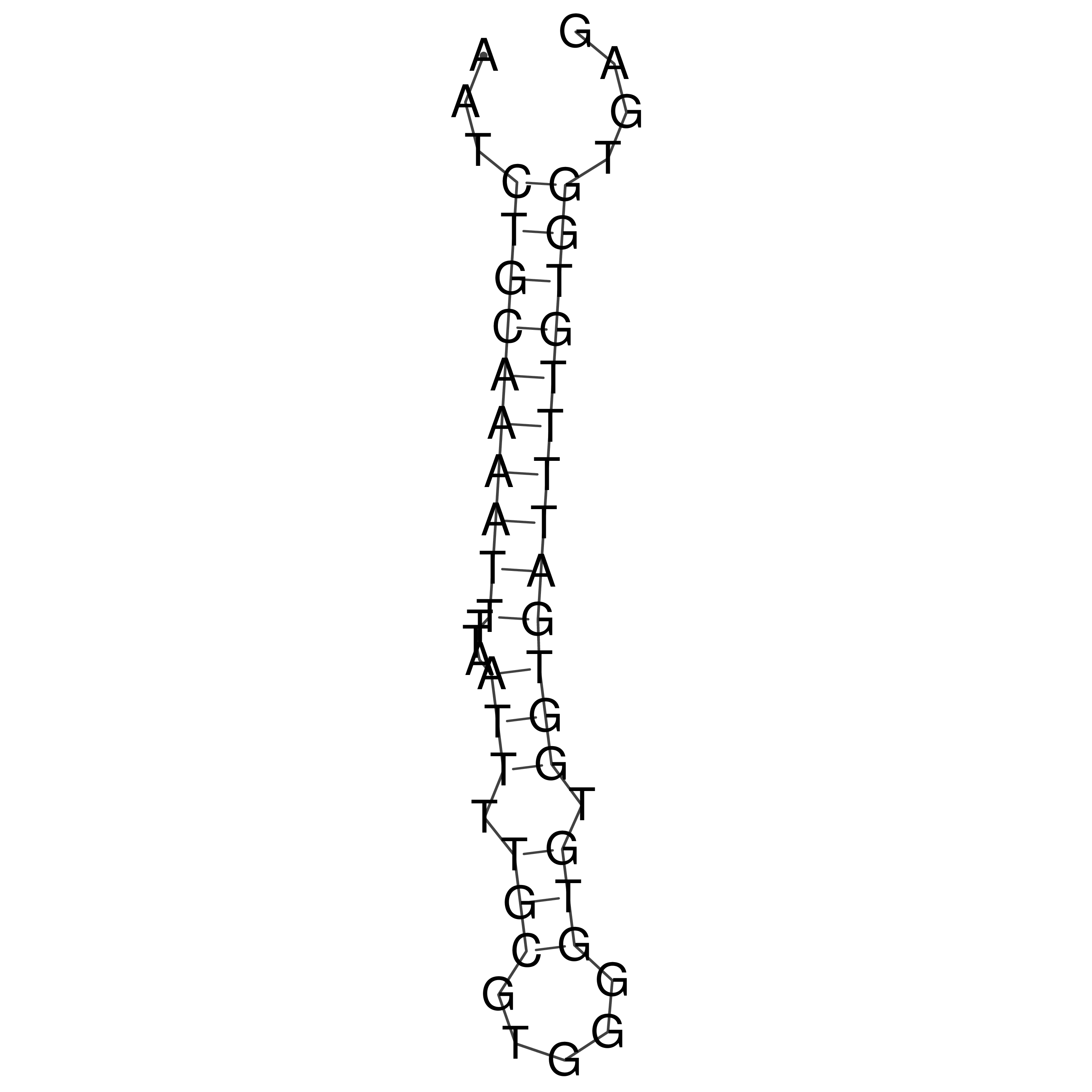
Relaxase
| ID | 1779 | GenBank | WP_025712711 |
| Name | traI_FTO35_RS30130_KZN_INI_KINF_24|unnamed45 |
UniProt ID | A0A7G9AB41 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190484.63 Da Isoelectric Point: 6.5078
>WP_025712711.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHKTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KVAHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIVQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGGSVVEFTPKQEKAIQ
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWAEQAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRAGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHKTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRHEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KVAHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIVQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGGSVVEFTPKQEKAIQ
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAITGGRVWAEQAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRAGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1362 | GenBank | WP_025712710 |
| Name | traD_FTO35_RS30125_KZN_INI_KINF_24|unnamed45 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85947.01 Da Isoelectric Point: 5.1165
>WP_025712710.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMRKDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMRKDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1436..30244
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FTO35_RS29935 (FTO35_29935) | 220..1041 | + | 822 | WP_023316395 | DUF932 domain-containing protein | - |
| FTO35_RS29940 (FTO35_29940) | 1074..1403 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| FTO35_RS29945 (FTO35_29945) | 1436..1921 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| FTO35_RS29950 (FTO35_29950) | 2353..2745 | + | 393 | WP_004194114 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| FTO35_RS29955 (FTO35_29955) | 2975..3676 | + | 702 | WP_025712700 | hypothetical protein | - |
| FTO35_RS29960 (FTO35_29960) | 3762..3962 | + | 201 | WP_046664192 | TraY domain-containing protein | - |
| FTO35_RS29965 (FTO35_29965) | 4030..4398 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| FTO35_RS29970 (FTO35_29970) | 4412..4717 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| FTO35_RS29975 (FTO35_29975) | 4737..5303 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| FTO35_RS29980 (FTO35_29980) | 5290..6030 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| FTO35_RS29985 (FTO35_29985) | 6030..7454 | + | 1425 | WP_015065626 | F-type conjugal transfer pilus assembly protein TraB | traB |
| FTO35_RS29990 (FTO35_29990) | 7528..8112 | + | 585 | WP_020277945 | type IV conjugative transfer system lipoprotein TraV | traV |
| FTO35_RS31835 | 8267..8665 | + | 399 | WP_020277946 | hypothetical protein | - |
| FTO35_RS31840 (FTO35_30000) | 8736..8984 | + | 249 | WP_223175793 | hypothetical protein | - |
| FTO35_RS30005 (FTO35_30005) | 9008..9244 | + | 237 | WP_042929429 | hypothetical protein | - |
| FTO35_RS30010 (FTO35_30010) | 9226..9537 | + | 312 | WP_025712701 | hypothetical protein | - |
| FTO35_RS30015 (FTO35_30015) | 9604..10008 | + | 405 | WP_004197817 | hypothetical protein | - |
| FTO35_RS30020 (FTO35_30020) | 10051..10350 | + | 300 | WP_025712702 | hypothetical protein | - |
| FTO35_RS30025 (FTO35_30025) | 10358..10756 | + | 399 | WP_074193813 | hypothetical protein | - |
| FTO35_RS30030 (FTO35_30030) | 10828..13467 | + | 2640 | WP_025712704 | type IV secretion system protein TraC | virb4 |
| FTO35_RS30035 (FTO35_30035) | 13467..13856 | + | 390 | WP_013214025 | type-F conjugative transfer system protein TrbI | - |
| FTO35_RS30040 (FTO35_30040) | 13856..14482 | + | 627 | WP_025712705 | type-F conjugative transfer system protein TraW | traW |
| FTO35_RS30045 (FTO35_30045) | 14526..15485 | + | 960 | WP_202560610 | conjugal transfer pilus assembly protein TraU | traU |
| FTO35_RS30050 (FTO35_30050) | 15498..16136 | + | 639 | WP_022644933 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| FTO35_RS30055 (FTO35_30055) | 16195..18150 | + | 1956 | WP_025712707 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| FTO35_RS30060 (FTO35_30060) | 18182..18436 | + | 255 | WP_020804203 | conjugal transfer protein TrbE | - |
| FTO35_RS30065 (FTO35_30065) | 18414..18662 | + | 249 | WP_004152675 | hypothetical protein | - |
| FTO35_RS30070 (FTO35_30070) | 18675..19001 | + | 327 | WP_004144402 | hypothetical protein | - |
| FTO35_RS30075 (FTO35_30075) | 19022..19774 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| FTO35_RS30080 (FTO35_30080) | 19785..20024 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| FTO35_RS30085 (FTO35_30085) | 19996..20553 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| FTO35_RS30090 (FTO35_30090) | 20599..21042 | + | 444 | WP_025712708 | F-type conjugal transfer protein TrbF | - |
| FTO35_RS30095 (FTO35_30095) | 21020..22399 | + | 1380 | WP_074187349 | conjugal transfer pilus assembly protein TraH | traH |
| FTO35_RS30100 (FTO35_30100) | 22399..25242 | + | 2844 | WP_020805032 | conjugal transfer mating-pair stabilization protein TraG | traG |
| FTO35_RS30105 (FTO35_30105) | 25253..25786 | + | 534 | WP_004197891 | hypothetical protein | - |
| FTO35_RS30115 (FTO35_30115) | 26194..26923 | + | 730 | Protein_35 | conjugal transfer complement resistance protein TraT | - |
| FTO35_RS31435 | 27116..27805 | + | 690 | WP_014343485 | hypothetical protein | - |
| FTO35_RS30125 (FTO35_30125) | 27932..30244 | + | 2313 | WP_025712710 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2788 | GenBank | NZ_VOOC01000099 |
| Plasmid name | KZN_INI_KINF_24|unnamed45 | Incompatibility group | IncFII |
| Plasmid size | 42130 bp | Coordinate of oriT [Strand] | 1994..2042 [-] |
| Host baterium | Klebsiella pneumoniae strain KZN_INI_KINF_24 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |