Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102332 |
| Name | oriT_KZN_INI_KINF_28|unnamed29 |
| Organism | Klebsiella pneumoniae strain KZN_INI_KINF_28 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_VOOE01000122 (23298..23347 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_KZN_INI_KINF_28|unnamed29
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file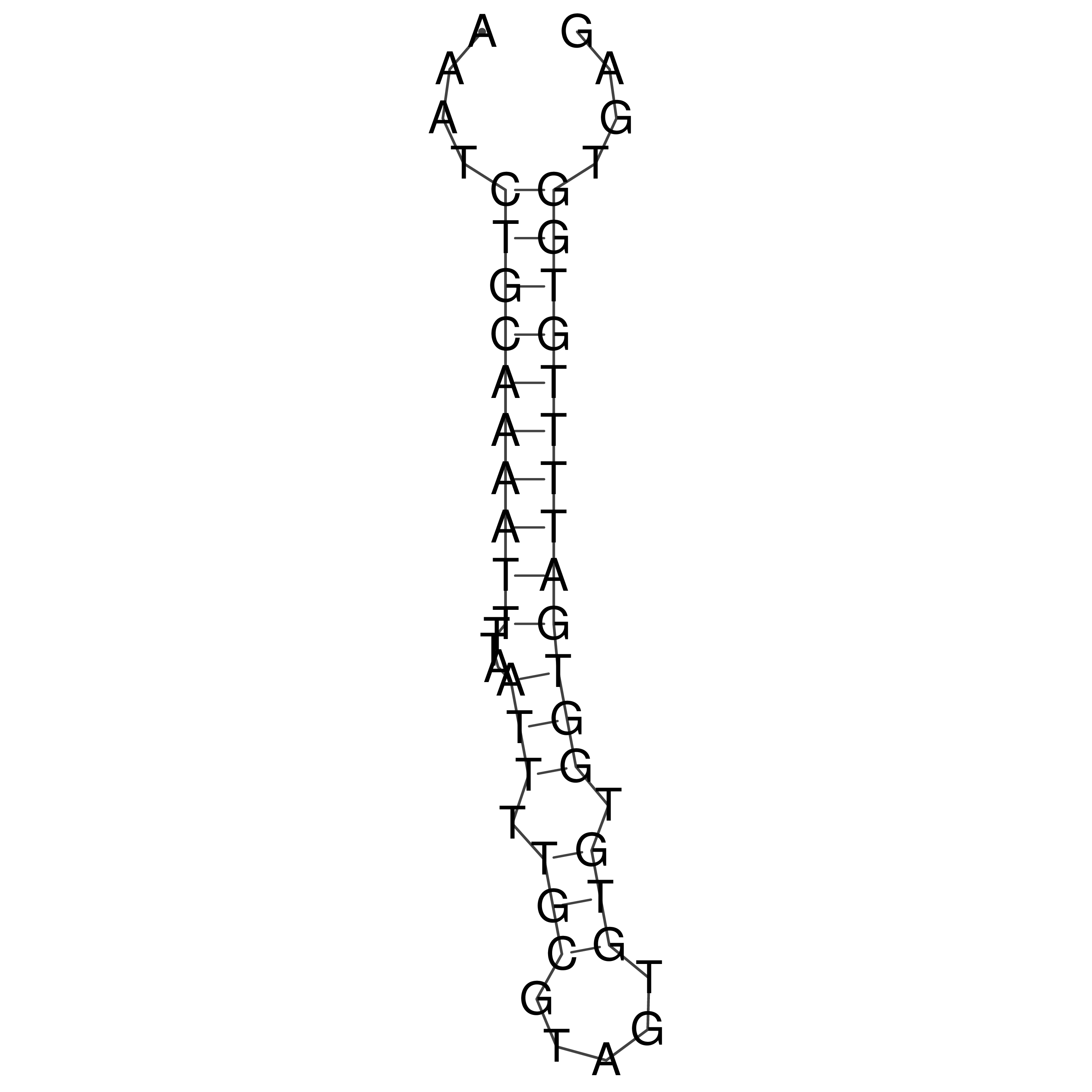
Relaxase
| ID | 1772 | GenBank | WP_015065542 |
| Name | traI_FTO37_RS30850_KZN_INI_KINF_28|unnamed29 |
UniProt ID | K7R3U7 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190318.03 Da Isoelectric Point: 6.2890
>WP_015065542.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMSREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALAVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDTAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGGGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMSREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALAVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDTAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGGGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1356 | GenBank | WP_015065541 |
| Name | traD_FTO37_RS30845_KZN_INI_KINF_28|unnamed29 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 86025.00 Da Isoelectric Point: 5.0577
>WP_015065541.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 22740..51707
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FTO37_RS30635 (FTO37_30645) | 18132..18359 | + | 228 | WP_001568050 | hypothetical protein | - |
| FTO37_RS30640 (FTO37_30650) | 18451..18681 | + | 231 | WP_009310031 | hypothetical protein | - |
| FTO37_RS30645 (FTO37_30655) | 18733..20088 | + | 1356 | WP_015065512 | DUF3560 domain-containing protein | - |
| FTO37_RS30650 (FTO37_30660) | 20136..20699 | + | 564 | WP_004152756 | methyltransferase | - |
| FTO37_RS30660 (FTO37_30670) | 21523..22345 | + | 823 | Protein_31 | DUF932 domain-containing protein | - |
| FTO37_RS30665 (FTO37_30675) | 22378..22707 | + | 330 | WP_015065524 | DUF5983 family protein | - |
| FTO37_RS30670 (FTO37_30680) | 22740..23225 | - | 486 | WP_015065525 | transglycosylase SLT domain-containing protein | virB1 |
| FTO37_RS30675 (FTO37_30685) | 23616..24032 | + | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| FTO37_RS30680 (FTO37_30690) | 24232..24951 | + | 720 | WP_015065526 | hypothetical protein | - |
| FTO37_RS30685 (FTO37_30695) | 25084..25287 | + | 204 | WP_171486139 | TraY domain-containing protein | - |
| FTO37_RS30690 (FTO37_30700) | 25352..25720 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| FTO37_RS30695 (FTO37_30705) | 25734..26039 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| FTO37_RS30700 (FTO37_30710) | 26059..26625 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| FTO37_RS30705 (FTO37_30715) | 26612..27352 | + | 741 | WP_015065528 | type-F conjugative transfer system secretin TraK | traK |
| FTO37_RS30710 (FTO37_30720) | 27352..28776 | + | 1425 | WP_004194260 | F-type conjugal transfer pilus assembly protein TraB | traB |
| FTO37_RS30715 (FTO37_30725) | 28890..29474 | + | 585 | WP_015065529 | type IV conjugative transfer system lipoprotein TraV | traV |
| FTO37_RS30720 (FTO37_30730) | 29606..30016 | + | 411 | WP_032329942 | hypothetical protein | - |
| FTO37_RS30725 (FTO37_30735) | 30122..30340 | + | 219 | WP_004152501 | hypothetical protein | - |
| FTO37_RS30730 (FTO37_30740) | 30341..30652 | + | 312 | WP_004152502 | hypothetical protein | - |
| FTO37_RS30735 (FTO37_30745) | 30719..31123 | + | 405 | WP_004152503 | hypothetical protein | - |
| FTO37_RS32420 | 31315..31557 | + | 243 | WP_004158946 | hypothetical protein | - |
| FTO37_RS30745 (FTO37_30755) | 31565..31963 | + | 399 | WP_011977783 | hypothetical protein | - |
| FTO37_RS30750 (FTO37_30760) | 32035..34674 | + | 2640 | WP_004152505 | type IV secretion system protein TraC | virb4 |
| FTO37_RS30755 (FTO37_30765) | 34674..35063 | + | 390 | WP_004152506 | type-F conjugative transfer system protein TrbI | - |
| FTO37_RS30760 (FTO37_30770) | 35063..35689 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| FTO37_RS30765 (FTO37_30775) | 35731..36120 | + | 390 | WP_015065532 | hypothetical protein | - |
| FTO37_RS30770 (FTO37_30780) | 36117..37106 | + | 990 | WP_011977785 | conjugal transfer pilus assembly protein TraU | traU |
| FTO37_RS30775 (FTO37_30785) | 37119..37757 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| FTO37_RS30780 (FTO37_30790) | 37816..39771 | + | 1956 | WP_030003482 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| FTO37_RS30785 (FTO37_30795) | 39803..40057 | + | 255 | WP_147015943 | conjugal transfer protein TrbE | - |
| FTO37_RS30790 (FTO37_30800) | 40035..40283 | + | 249 | WP_004152675 | hypothetical protein | - |
| FTO37_RS30795 (FTO37_30805) | 40296..40622 | + | 327 | WP_004144402 | hypothetical protein | - |
| FTO37_RS30800 (FTO37_30810) | 40643..41395 | + | 753 | WP_020803149 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| FTO37_RS30805 (FTO37_30815) | 41406..41645 | + | 240 | WP_004152687 | type-F conjugative transfer system pilin chaperone TraQ | - |
| FTO37_RS30810 (FTO37_30820) | 41617..42189 | + | 573 | WP_023292147 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| FTO37_RS30815 (FTO37_30825) | 42182..42610 | + | 429 | WP_015065560 | hypothetical protein | - |
| FTO37_RS30820 (FTO37_30830) | 42597..43967 | + | 1371 | WP_004159274 | conjugal transfer pilus assembly protein TraH | traH |
| FTO37_RS30825 (FTO37_30835) | 43967..46810 | + | 2844 | WP_015065537 | conjugal transfer mating-pair stabilization protein TraG | traG |
| FTO37_RS30830 (FTO37_30840) | 46821..47345 | + | 525 | WP_015065538 | hypothetical protein | - |
| FTO37_RS30835 (FTO37_30845) | 47654..48385 | + | 732 | WP_032329926 | conjugal transfer complement resistance protein TraT | - |
| FTO37_RS31895 | 48577..49266 | + | 690 | WP_015065540 | hypothetical protein | - |
| FTO37_RS30845 (FTO37_30855) | 49395..51707 | + | 2313 | WP_015065541 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2776 | GenBank | NZ_VOOE01000122 |
| Plasmid name | KZN_INI_KINF_28|unnamed29 | Incompatibility group | IncFII |
| Plasmid size | 63931 bp | Coordinate of oriT [Strand] | 23298..23347 [-] |
| Host baterium | Klebsiella pneumoniae strain KZN_INI_KINF_28 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |