Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102326 |
| Name | oriT_p7209-49 |
| Organism | Serratia marcescens strain 7209 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_PEHC01000009 (1481..1586 [+], 106 nt) |
| oriT length | 106 nt |
| IRs (inverted repeats) | 30..35, 41..46 (CCGCCG..CGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 106 nt
>oriT_p7209-49
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file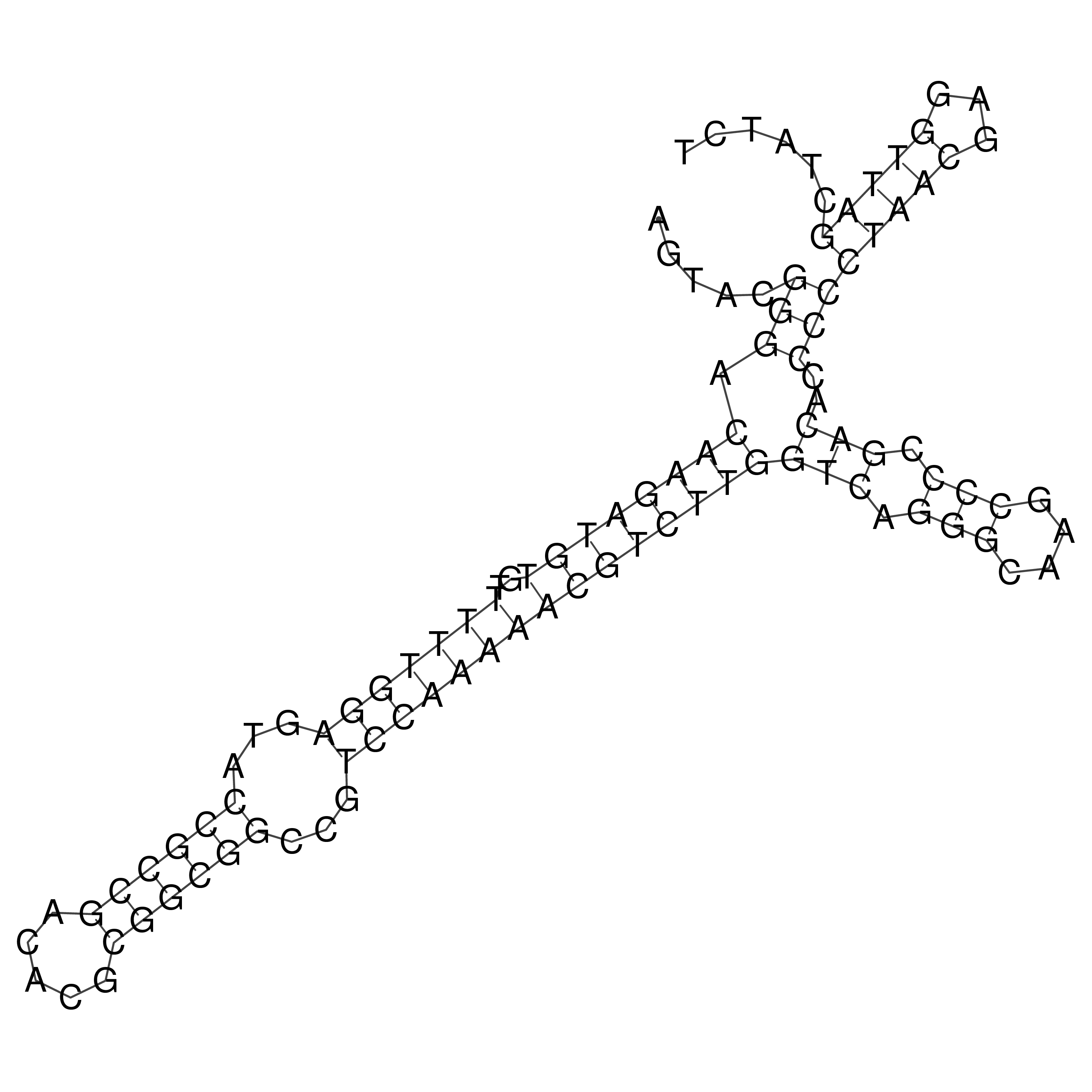
Relaxase
| ID | 1770 | GenBank | WP_101453311 |
| Name | Relaxase_CS371_RS14350_p7209-49 |
UniProt ID | _ |
| Length | 659 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 659 a.a. Molecular weight: 75193.92 Da Isoelectric Point: 9.9948
>WP_101453311.1 TraI/MobA(P) family conjugative relaxase [Serratia marcescens]
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAALNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQFRSESDDKFNPK
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAALNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQFRSESDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1355 | GenBank | WP_101453316 |
| Name | t4cp2_CS371_RS14465_p7209-49 |
UniProt ID | _ |
| Length | 695 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 695 a.a. Molecular weight: 79539.85 Da Isoelectric Point: 4.7667
>WP_101453316.1 conjugal transfer protein TrbC [Serratia marcescens]
MQQESVDSKKVLRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYERARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFPEDSGDIRMDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGK
KEEVLLTHALNNQAPYGLIYDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANT
RVKYFESIEDRKTFEILRETVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQG
VISFQDALVRSAAFYIPDNEKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMRFPPIK
LFGYDKAANSLLEKIEVFYELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
MQQESVDSKKVLRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYERARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFPEDSGDIRMDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGK
KEEVLLTHALNNQAPYGLIYDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANT
RVKYFESIEDRKTFEILRETVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQG
VISFQDALVRSAAFYIPDNEKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMRFPPIK
LFGYDKAANSLLEKIEVFYELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4382..31681
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CS371_RS14335 (CS371_14190) | 1..446 | + | 446 | WP_101453310 | hypothetical protein | - |
| CS371_RS14340 (CS371_14195) | 948..1343 | - | 396 | WP_019725163 | hypothetical protein | - |
| CS371_RS28675 | 1378..1905 | + | 528 | WP_172740549 | plasmid mobilization protein MobA | - |
| CS371_RS14350 (CS371_14205) | 1892..3871 | + | 1980 | WP_101453311 | TraI/MobA(P) family conjugative relaxase | - |
| CS371_RS14355 (CS371_14210) | 3885..4385 | + | 501 | WP_004187320 | DotD/TraH family lipoprotein | - |
| CS371_RS14360 (CS371_14215) | 4382..5161 | + | 780 | WP_004187315 | type IV secretory system conjugative DNA transfer family protein | traI |
| CS371_RS14365 (CS371_14220) | 5172..6335 | + | 1164 | WP_004187313 | plasmid transfer ATPase TraJ | virB11 |
| CS371_RS14370 (CS371_14225) | 6325..6585 | + | 261 | WP_004187310 | IcmT/TraK family protein | traK |
| CS371_RS28680 | 6610..9813 | + | 3204 | WP_242490474 | LPD7 domain-containing protein | - |
| CS371_RS27975 (CS371_14235) | 9968..10480 | + | 513 | WP_011091071 | hypothetical protein | traL |
| CS371_RS14380 (CS371_14240) | 10481..10693 | - | 213 | WP_305953721 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| CS371_RS14385 (CS371_14245) | 10665..11075 | - | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| CS371_RS14390 (CS371_14250) | 11143..11856 | + | 714 | WP_004187467 | DotI/IcmL family type IV secretion protein | traM |
| CS371_RS14400 (CS371_14255) | 11865..13016 | + | 1152 | WP_004187471 | DotH/IcmK family type IV secretion protein | traN |
| CS371_RS28685 | 13028..13225 | + | 198 | WP_242490475 | hypothetical protein | traO |
| CS371_RS14405 (CS371_14260) | 13229..14377 | + | 1149 | WP_277949772 | conjugal transfer protein TraO | traO |
| CS371_RS14410 (CS371_14265) | 14389..15093 | + | 705 | WP_015060002 | conjugal transfer protein TraP | traP |
| CS371_RS14415 (CS371_14270) | 15117..15647 | + | 531 | WP_004187478 | conjugal transfer protein TraQ | traQ |
| CS371_RS14420 (CS371_14275) | 15664..16053 | + | 390 | WP_004187479 | DUF6750 family protein | traR |
| CS371_RS14425 (CS371_14280) | 16099..16593 | + | 495 | WP_004187480 | hypothetical protein | - |
| CS371_RS14430 (CS371_14285) | 16590..19640 | + | 3051 | WP_101453313 | conjugal transfer protein | traU |
| CS371_RS14435 (CS371_14290) | 19637..20845 | + | 1209 | WP_101453314 | conjugal transfer protein TraW | traW |
| CS371_RS14440 (CS371_14295) | 20842..21438 | + | 597 | WP_015060003 | hypothetical protein | - |
| CS371_RS28960 | 21431..22735 | + | 1305 | WP_277949771 | DotA/TraY family protein | traY |
| CS371_RS28965 | 22722..23627 | + | 906 | WP_277949773 | DotA/TraY family protein | traY |
| CS371_RS14450 (CS371_14305) | 23629..24282 | + | 654 | WP_015060005 | hypothetical protein | - |
| CS371_RS14455 (CS371_14310) | 24351..24593 | + | 243 | WP_023893611 | IncL/M type plasmid replication protein RepC | - |
| CS371_RS14460 (CS371_14315) | 24889..25944 | + | 1056 | WP_015060006 | plasmid replication initiator RepA | - |
| CS371_RS28970 | 27167..27262 | + | 96 | WP_004206884 | DinQ-like type I toxin DqlB | - |
| CS371_RS14465 (CS371_14320) | 27313..29400 | - | 2088 | WP_101453316 | conjugal transfer protein TrbC | - |
| CS371_RS14470 (CS371_14325) | 29413..30363 | - | 951 | WP_004206886 | DsbC family protein | trbB |
| CS371_RS14475 (CS371_14330) | 30374..31681 | - | 1308 | WP_242490476 | conjugal transfer protein TrbA | trbA |
| CS371_RS14480 (CS371_14335) | 31681..32076 | - | 396 | WP_004187436 | lytic transglycosylase domain-containing protein | - |
| CS371_RS14485 (CS371_14340) | 32181..32564 | - | 384 | WP_019725042 | DUF1496 domain-containing protein | - |
| CS371_RS14490 (CS371_14345) | 32644..33297 | + | 654 | WP_004206890 | type II CAAX endopeptidase family protein | - |
| CS371_RS27980 | 33389..33646 | + | 258 | WP_004187429 | type II toxin-antitoxin system antitoxin PemI | - |
| CS371_RS14495 (CS371_14350) | 33648..33980 | + | 333 | WP_144236188 | type II toxin-antitoxin system toxin endoribonuclease PemK-mt | - |
| CS371_RS14500 (CS371_14355) | 34073..34507 | + | 435 | WP_004187425 | translesion error-prone DNA polymerase V autoproteolytic subunit | - |
| CS371_RS14505 (CS371_14360) | 34495..35762 | + | 1268 | Protein_38 | translesion error-prone DNA polymerase V subunit UmuC | - |
| CS371_RS14510 (CS371_14365) | 35885..36094 | - | 210 | WP_004187413 | hypothetical protein | - |
| CS371_RS28690 | 36230..36652 | - | 423 | WP_242490477 | hypothetical protein | - |
Host bacterium
| ID | 2770 | GenBank | NZ_PEHC01000009 |
| Plasmid name | p7209-49 | Incompatibility group | IncL/M |
| Plasmid size | 48903 bp | Coordinate of oriT [Strand] | 1481..1586 [+] |
| Host baterium | Serratia marcescens strain 7209 |
Cargo genes
| Drug resistance gene | blaTEM-1C, blaCTX-M-15 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |