Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102278 |
| Name | oriT_DSM 25444|unnamed3 |
| Organism | Klebsiella michiganensis strain DSM 25444 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_PRDB01000032 (84817..84864 [-], 48 nt) |
| oriT length | 48 nt |
| IRs (inverted repeats) | 5..12, 15..22 (GCAAAATT..AATTTTGC) |
| Location of nic site | 31..32 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 48 nt
>oriT_DSM 25444|unnamed3
ATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
ATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file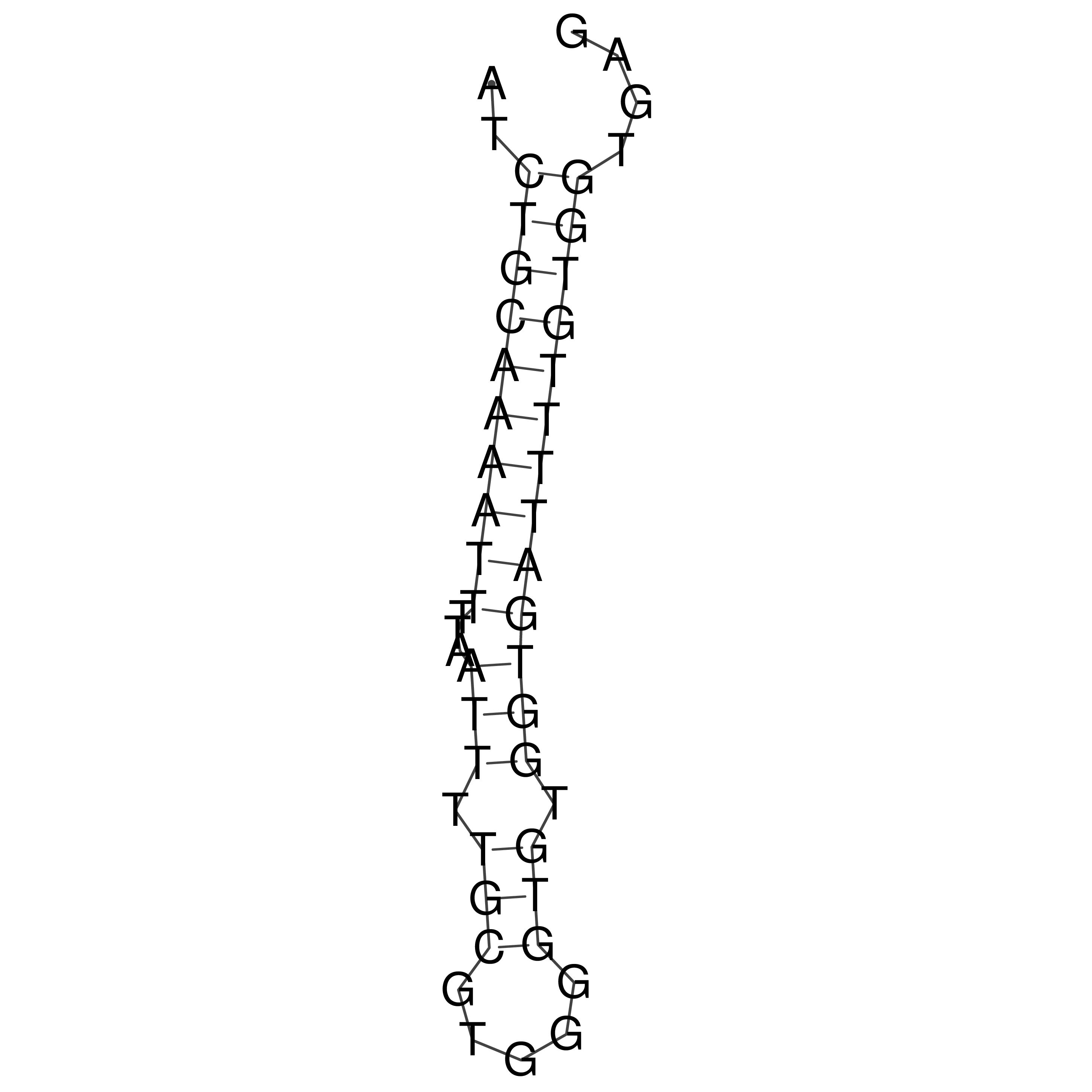
Relaxase
| ID | 1750 | GenBank | WP_064362890 |
| Name | traI_CDA56_RS30485_DSM 25444|unnamed3 |
UniProt ID | A0A443W956 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190128.87 Da Isoelectric Point: 6.1737
>WP_064362890.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MMSIGSVKSAGSASNYYTDKDNYYVIGSMEERWQGKGAEALGLDGKAVDKAVFTELLKGKLPDGSDLTRM
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTVAVKQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQTAFGKLYREALKPLVE
KLGYETKVVGKHGMWEMKDVPVEAFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADAHAAELARAPVAPEKTDGPDITDVVTRAIAGLSDRKVQFTYADVLARTVGQLE
AKPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDAGVVREKP
FSDAVSVLAQDRPAMGIVAGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSREFLAGDARLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGKQATADIISEPDKSARYSRLAQEFAASVREGQESVAQISGTREQNVLNNVIRDTLKNEGALGNRD
MAVTALTPVWLDSKSRGVRDFYREGMVMERWDPEARKHDRFVIDRVTASSNMLTLKDREGERLDLKVSAV
DSQWTLFRTDKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQTLPVGSSPFDGI
KVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGNHIRLYSAQDAARTTEKLARHTAFSVVSE
QLKARSGEADLEAAIAQQKAGLHTPAEQAVHLSIPLLESNSLTFTRPQLLAAALETGDGKVPMKDIEATI
QAQIKAGSLLNVPVASGHGNDLLISRQTWDAEKSVLTRVLEGKDAVAPLMGRVPGSLMTDLTAGQRAATR
MILETPDRFTVVQGYAGVGKTTQFKAVMSAISLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLH
DTQLQQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDISRALSTIEQVTPEQVPRKDGAWVPENSVTEFTPAQEKAIQ
DALKEGKEIPPGTPVSLHDAIVKDYTGRTPAAQASTLIITHLNADRRALNGLIHDARVQNGEVSKEEVTL
PVLVTSNIRDGELRKLSTWSEHQGAVALLDNTYQRIESVDKENQLITLKDNEGNIRLLSPREASAEGVTL
YRPEHITVSQGDRMRFSKSDPERGYVANSVWEVKTVSGDSVTLSDGKTTRTLNPKADEAQRHIDLAYAIT
AHGAQGASEPFAIALEGVEGRRKALVSFESAYVALSRMKQHAQVYTDNREGWTTAVSRSQEKASAHDVLE
PQNARAVKNAGQLYGRAQPLDETAAGRAALQQSGLARGRSPGKFISPGKKYPQPHVALPAFDKNGKEAGI
WLSPLTDRDGRLEAIGGEGRIMGDETARFVALQSSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDR
PWNPEAITGGRIWADPLPNPPLPDTGTDIPLPPEVLAQRAAEEAQRREMEKQAEQATREVAGEEKKAGEP
GERIKEVIGDVIRGLERDRPGEEKTRLPDDPQTRRQEAAIQQVASESLQRERLQQMERDMVRDLNREKTL
GGD
MMSIGSVKSAGSASNYYTDKDNYYVIGSMEERWQGKGAEALGLDGKAVDKAVFTELLKGKLPDGSDLTRM
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTVAVKQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQTAFGKLYREALKPLVE
KLGYETKVVGKHGMWEMKDVPVEAFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADAHAAELARAPVAPEKTDGPDITDVVTRAIAGLSDRKVQFTYADVLARTVGQLE
AKPGVFELARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDAGVVREKP
FSDAVSVLAQDRPAMGIVAGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSREFLAGDARLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGKQATADIISEPDKSARYSRLAQEFAASVREGQESVAQISGTREQNVLNNVIRDTLKNEGALGNRD
MAVTALTPVWLDSKSRGVRDFYREGMVMERWDPEARKHDRFVIDRVTASSNMLTLKDREGERLDLKVSAV
DSQWTLFRTDKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQTLPVGSSPFDGI
KVGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGNHIRLYSAQDAARTTEKLARHTAFSVVSE
QLKARSGEADLEAAIAQQKAGLHTPAEQAVHLSIPLLESNSLTFTRPQLLAAALETGDGKVPMKDIEATI
QAQIKAGSLLNVPVASGHGNDLLISRQTWDAEKSVLTRVLEGKDAVAPLMGRVPGSLMTDLTAGQRAATR
MILETPDRFTVVQGYAGVGKTTQFKAVMSAISLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLH
DTQLQQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQR
SAADVAIMKEIVRQVPELRPAVYSLIDRDISRALSTIEQVTPEQVPRKDGAWVPENSVTEFTPAQEKAIQ
DALKEGKEIPPGTPVSLHDAIVKDYTGRTPAAQASTLIITHLNADRRALNGLIHDARVQNGEVSKEEVTL
PVLVTSNIRDGELRKLSTWSEHQGAVALLDNTYQRIESVDKENQLITLKDNEGNIRLLSPREASAEGVTL
YRPEHITVSQGDRMRFSKSDPERGYVANSVWEVKTVSGDSVTLSDGKTTRTLNPKADEAQRHIDLAYAIT
AHGAQGASEPFAIALEGVEGRRKALVSFESAYVALSRMKQHAQVYTDNREGWTTAVSRSQEKASAHDVLE
PQNARAVKNAGQLYGRAQPLDETAAGRAALQQSGLARGRSPGKFISPGKKYPQPHVALPAFDKNGKEAGI
WLSPLTDRDGRLEAIGGEGRIMGDETARFVALQSSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDR
PWNPEAITGGRIWADPLPNPPLPDTGTDIPLPPEVLAQRAAEEAQRREMEKQAEQATREVAGEEKKAGEP
GERIKEVIGDVIRGLERDRPGEEKTRLPDDPQTRRQEAAIQQVASESLQRERLQQMERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1339 | GenBank | WP_064174013 |
| Name | traD_CDA56_RS30480_DSM 25444|unnamed3 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 85728.45 Da Isoelectric Point: 4.8494
>WP_064174013.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRTLDTRVDARLSALLEAREAEGSLARALFTPDAPEPEQSDADSRTSPGQPEPESAP
VSPAPVKSASTAETLAAQPSVKASEAPQAKVTTVPLTRPVPAATTTAAASATGSSAAATATTGGTEQELA
QQPAEQGQEMMPAGMNEDGEIEDMQAYDAWLADEQTQREMQRREEVNINHSHRRDEQDDIEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRTLDTRVDARLSALLEAREAEGSLARALFTPDAPEPEQSDADSRTSPGQPEPESAP
VSPAPVKSASTAETLAAQPSVKASEAPQAKVTTVPLTRPVPAATTTAAASATGSSAAATATTGGTEQELA
QQPAEQGQEMMPAGMNEDGEIEDMQAYDAWLADEQTQREMQRREEVNINHSHRRDEQDDIEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 84259..110829
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CDA56_RS30255 (CDA56_30250) | 79287..79637 | + | 351 | WP_004152758 | hypothetical protein | - |
| CDA56_RS30270 (CDA56_30265) | 80266..80622 | + | 357 | WP_019706019 | hypothetical protein | - |
| CDA56_RS30275 (CDA56_30270) | 80683..80895 | + | 213 | WP_019706020 | hypothetical protein | - |
| CDA56_RS30280 (CDA56_30275) | 80906..81130 | + | 225 | WP_014343499 | hypothetical protein | - |
| CDA56_RS30285 (CDA56_30280) | 81211..81531 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| CDA56_RS30290 (CDA56_30285) | 81521..81799 | + | 279 | WP_032411288 | helix-turn-helix transcriptional regulator | - |
| CDA56_RS30295 (CDA56_30290) | 81800..82213 | + | 414 | WP_032411285 | helix-turn-helix domain-containing protein | - |
| CDA56_RS30310 (CDA56_30305) | 83043..83864 | + | 822 | WP_004182076 | DUF932 domain-containing protein | - |
| CDA56_RS30315 (CDA56_30310) | 83897..84226 | + | 330 | WP_032420969 | DUF5983 family protein | - |
| CDA56_RS30320 (CDA56_30315) | 84259..84744 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| CDA56_RS30325 (CDA56_30320) | 85165..85560 | + | 396 | WP_022644714 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| CDA56_RS30330 (CDA56_30325) | 85732..86418 | + | 687 | WP_022644713 | transcriptional regulator TraJ family protein | - |
| CDA56_RS30335 (CDA56_30330) | 86497..86877 | + | 381 | WP_032409797 | TraY domain-containing protein | - |
| CDA56_RS30340 (CDA56_30335) | 86936..87301 | + | 366 | WP_020314752 | type IV conjugative transfer system pilin TraA | - |
| CDA56_RS30345 (CDA56_30340) | 87315..87620 | + | 306 | WP_020323523 | type IV conjugative transfer system protein TraL | traL |
| CDA56_RS30350 (CDA56_30345) | 87640..88206 | + | 567 | WP_064373744 | type IV conjugative transfer system protein TraE | traE |
| CDA56_RS30355 (CDA56_30350) | 88193..88933 | + | 741 | WP_064362918 | type-F conjugative transfer system secretin TraK | traK |
| CDA56_RS30360 (CDA56_30355) | 88933..90348 | + | 1416 | WP_064362917 | F-type conjugal transfer pilus assembly protein TraB | traB |
| CDA56_RS30815 | 90341..90943 | + | 603 | WP_115723427 | conjugal transfer protein TraP | - |
| CDA56_RS31625 (CDA56_30365) | 91011..91148 | + | 138 | Protein_111 | conjugal transfer protein TraP | - |
| CDA56_RS30375 (CDA56_30370) | 91171..91734 | + | 564 | WP_064362913 | type IV conjugative transfer system lipoprotein TraV | traV |
| CDA56_RS30380 (CDA56_30375) | 91881..92084 | + | 204 | WP_064362911 | hypothetical protein | - |
| CDA56_RS31115 | 92325..92462 | + | 138 | WP_164879260 | hypothetical protein | - |
| CDA56_RS31120 | 92497..92739 | + | 243 | WP_074181267 | hypothetical protein | - |
| CDA56_RS30390 (CDA56_30385) | 92768..93067 | + | 300 | WP_020315144 | hypothetical protein | - |
| CDA56_RS30395 (CDA56_30390) | 93084..93308 | + | 225 | WP_022644711 | hypothetical protein | - |
| CDA56_RS30400 (CDA56_30395) | 93305..93709 | + | 405 | WP_064362909 | hypothetical protein | - |
| CDA56_RS30405 (CDA56_30400) | 93842..96481 | + | 2640 | WP_064362907 | type IV secretion system protein TraC | virb4 |
| CDA56_RS30410 (CDA56_30405) | 96514..96915 | + | 402 | WP_064362936 | type-F conjugative transfer system protein TrbI | - |
| CDA56_RS30415 (CDA56_30410) | 96912..97538 | + | 627 | WP_064362905 | type-F conjugative transfer system protein TraW | traW |
| CDA56_RS30420 (CDA56_30415) | 97558..98541 | + | 984 | WP_155553410 | conjugal transfer pilus assembly protein TraU | traU |
| CDA56_RS30425 (CDA56_30420) | 98554..99171 | + | 618 | WP_064362903 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| CDA56_RS30430 (CDA56_30425) | 99168..101048 | + | 1881 | WP_064362901 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| CDA56_RS30435 (CDA56_30430) | 101086..101367 | + | 282 | WP_064174416 | hypothetical protein | - |
| CDA56_RS30440 (CDA56_30435) | 101357..101584 | + | 228 | WP_064174417 | conjugal transfer protein TrbE | - |
| CDA56_RS30445 (CDA56_30440) | 101595..101921 | + | 327 | WP_064362899 | hypothetical protein | - |
| CDA56_RS30450 (CDA56_30445) | 101940..102692 | + | 753 | WP_040118488 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| CDA56_RS30455 (CDA56_30450) | 102703..102942 | + | 240 | WP_047722563 | type-F conjugative transfer system pilin chaperone TraQ | - |
| CDA56_RS30460 (CDA56_30455) | 102914..103492 | + | 579 | WP_064362897 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| CDA56_RS30465 (CDA56_30460) | 103479..104849 | + | 1371 | WP_064362895 | conjugal transfer pilus assembly protein TraH | traH |
| CDA56_RS30470 (CDA56_30465) | 104849..107719 | + | 2871 | WP_064362893 | conjugal transfer mating-pair stabilization protein TraG | traG |
| CDA56_RS30475 (CDA56_30470) | 107853..108293 | + | 441 | WP_224313869 | conjugal transfer protein TraS | - |
| CDA56_RS30480 (CDA56_30475) | 108529..110829 | + | 2301 | WP_064174013 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2722 | GenBank | NZ_PRDB01000032 |
| Plasmid name | DSM 25444|unnamed3 | Incompatibility group | IncFII |
| Plasmid size | 136210 bp | Coordinate of oriT [Strand] | 84817..84864 [-] |
| Host baterium | Klebsiella michiganensis strain DSM 25444 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | ncrY, ncrC, ncrB, ncrA, chrA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |