Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102248 |
| Name | oriT_pB16 P26 |
| Organism | Klebsiella pneumoniae strain B16 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_NTCW01000077 (49356..49405 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pB16 P26
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file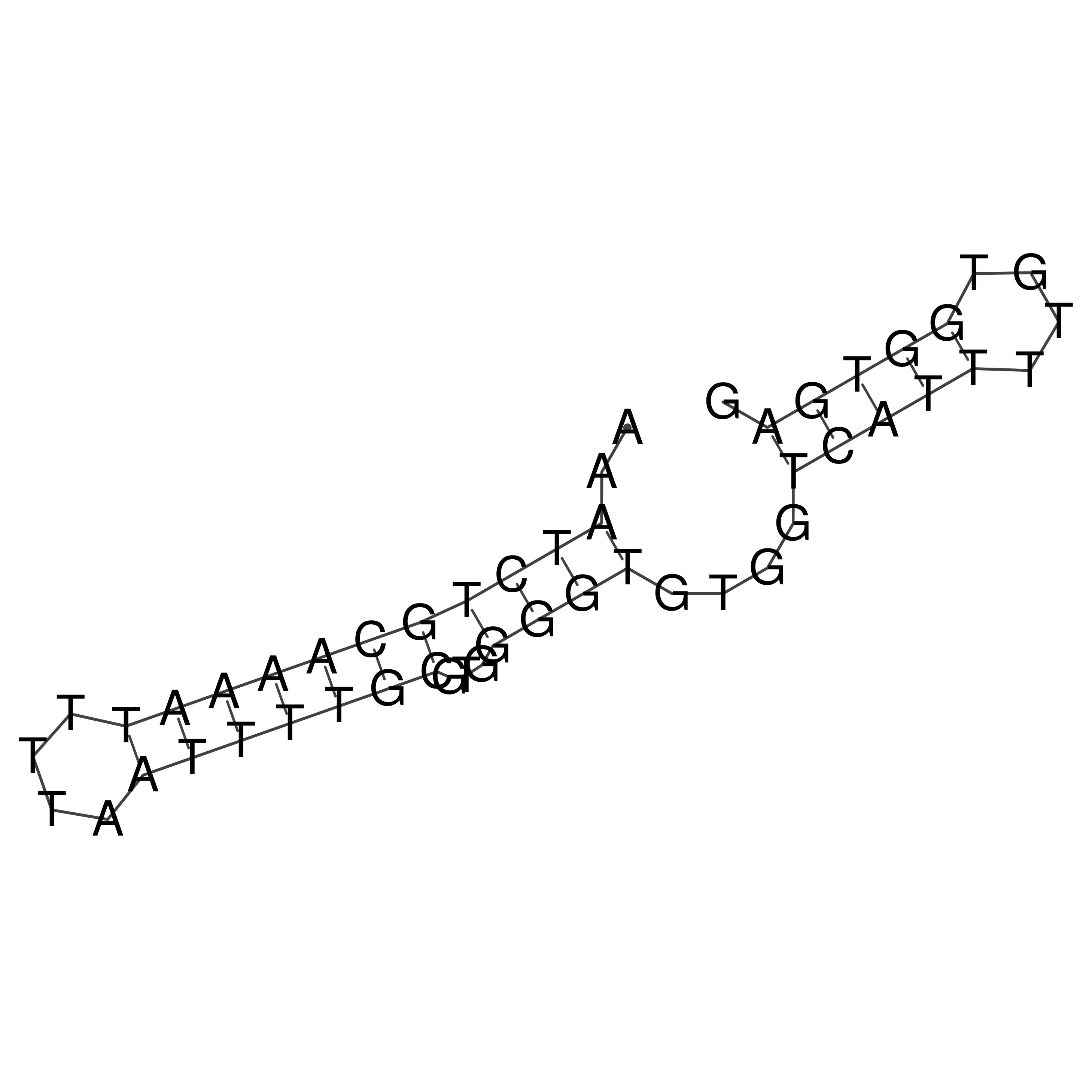
Relaxase
| ID | 1730 | GenBank | WP_065898399 |
| Name | traI_CLI87_RS28780_pB16 P26 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190718.76 Da Isoelectric Point: 6.2077
>WP_065898399.1 conjugative transfer relaxase/helicase TraI [Klebsiella pneumoniae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1313 | GenBank | WP_004152630 |
| Name | traD_CLI87_RS28785_pB16 P26 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85951.02 Da Isoelectric Point: 5.0632
>WP_004152630.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRRLDTRVDARLSALLEEREAEGSLARALFTPDAPASGPADTDSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAAEPSVRATEPPVLRVTTVPLIKPKAAAAAAAASTASSAGIPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRRLDTRVDARLSALLEEREAEGSLARALFTPDAPASGPADTDSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAAEPSVRATEPPVLRVTTVPLIKPKAAAAAAAASTASSAGIPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 21545..49963
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CLI87_RS28785 (CLI87_28795) | 21545..23854 | - | 2310 | WP_004152630 | type IV conjugative transfer system coupling protein TraD | virb4 |
| CLI87_RS28790 (CLI87_28800) | 24214..24945 | - | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| CLI87_RS28795 (CLI87_28805) | 25135..25662 | - | 528 | WP_004153030 | hypothetical protein | - |
| CLI87_RS28800 (CLI87_28810) | 25668..28517 | - | 2850 | WP_004178054 | conjugal transfer mating-pair stabilization protein TraG | traG |
| CLI87_RS28805 (CLI87_28815) | 28517..29887 | - | 1371 | WP_004178055 | conjugal transfer pilus assembly protein TraH | traH |
| CLI87_RS28810 (CLI87_28820) | 29874..30302 | - | 429 | WP_004152689 | hypothetical protein | - |
| CLI87_RS28815 (CLI87_28825) | 30295..30867 | - | 573 | WP_004152688 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| CLI87_RS28820 (CLI87_28830) | 30839..31078 | - | 240 | WP_004152687 | type-F conjugative transfer system pilin chaperone TraQ | - |
| CLI87_RS28825 (CLI87_28835) | 31089..31841 | - | 753 | WP_004152686 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| CLI87_RS28830 (CLI87_28840) | 31862..32188 | - | 327 | WP_004152685 | hypothetical protein | - |
| CLI87_RS32465 (CLI87_28845) | 32234..32419 | - | 186 | WP_004152684 | hypothetical protein | - |
| CLI87_RS28840 (CLI87_28850) | 32416..32652 | - | 237 | WP_004152683 | conjugal transfer protein TrbE | - |
| CLI87_RS28845 (CLI87_28855) | 32684..34639 | - | 1956 | WP_004153093 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| CLI87_RS28850 (CLI87_28860) | 34687..35325 | - | 639 | WP_004152682 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| CLI87_RS28855 (CLI87_28865) | 35338..36297 | - | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| CLI87_RS28860 (CLI87_28870) | 36341..36967 | - | 627 | WP_094387712 | type-F conjugative transfer system protein TraW | traW |
| CLI87_RS28865 (CLI87_28875) | 36967..37356 | - | 390 | WP_004152591 | type-F conjugative transfer system protein TrbI | - |
| CLI87_RS28870 (CLI87_28880) | 37356..39995 | - | 2640 | WP_004152592 | type IV secretion system protein TraC | virb4 |
| CLI87_RS28875 (CLI87_28885) | 40067..40465 | - | 399 | WP_004153071 | hypothetical protein | - |
| CLI87_RS28885 (CLI87_28895) | 40761..41165 | - | 405 | WP_004152594 | hypothetical protein | - |
| CLI87_RS28890 (CLI87_28900) | 41232..41549 | - | 318 | WP_004152595 | hypothetical protein | - |
| CLI87_RS28895 (CLI87_28905) | 41550..41768 | - | 219 | WP_004171484 | hypothetical protein | - |
| CLI87_RS28900 (CLI87_28910) | 41792..42082 | - | 291 | WP_004152596 | hypothetical protein | - |
| CLI87_RS28905 (CLI87_28915) | 42087..42497 | - | 411 | WP_004152597 | lipase chaperone | - |
| CLI87_RS28910 (CLI87_28920) | 42629..43198 | - | 570 | WP_004152598 | type IV conjugative transfer system lipoprotein TraV | traV |
| CLI87_RS28915 (CLI87_28925) | 43221..43817 | - | 597 | WP_004152599 | conjugal transfer pilus-stabilizing protein TraP | - |
| CLI87_RS28920 (CLI87_28930) | 43810..45234 | - | 1425 | WP_004152600 | F-type conjugal transfer pilus assembly protein TraB | traB |
| CLI87_RS28925 (CLI87_28935) | 45234..45974 | - | 741 | WP_004152601 | type-F conjugative transfer system secretin TraK | traK |
| CLI87_RS28930 (CLI87_28940) | 45961..46527 | - | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| CLI87_RS28935 (CLI87_28945) | 46547..46852 | - | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| CLI87_RS28940 (CLI87_28950) | 46866..47234 | - | 369 | WP_004178060 | type IV conjugative transfer system pilin TraA | - |
| CLI87_RS28945 (CLI87_28955) | 47288..47659 | - | 372 | WP_004208838 | TraY domain-containing protein | - |
| CLI87_RS28950 (CLI87_28960) | 47743..48438 | - | 696 | WP_004178061 | transcriptional regulator TraJ family protein | - |
| CLI87_RS28955 (CLI87_28965) | 48652..49044 | - | 393 | WP_004178062 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| CLI87_RS28960 (CLI87_28970) | 49478..49963 | + | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| CLI87_RS28965 (CLI87_28975) | 49996..50325 | - | 330 | WP_011977736 | DUF5983 family protein | - |
| CLI87_RS28970 (CLI87_28980) | 50358..51179 | - | 822 | WP_004178064 | DUF932 domain-containing protein | - |
| CLI87_RS28985 (CLI87_28995) | 51809..52159 | - | 351 | WP_004152758 | hypothetical protein | - |
| CLI87_RS28990 (CLI87_29000) | 52156..52428 | - | 273 | WP_004152759 | hypothetical protein | - |
| CLI87_RS28995 (CLI87_29005) | 53128..53289 | - | 162 | WP_004118124 | type I toxin-antitoxin system Hok family toxin | - |
| CLI87_RS29000 (CLI87_29010) | 53361..54440 | + | 1080 | WP_004220208 | IS481-like element ISKpn28 family transposase | - |
Host bacterium
| ID | 2692 | GenBank | NZ_NTCW01000077 |
| Plasmid name | pB16 P26 | Incompatibility group | IncFII |
| Plasmid size | 65332 bp | Coordinate of oriT [Strand] | 49356..49405 [+] |
| Host baterium | Klebsiella pneumoniae strain B16 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |