Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102137 |
| Name | oriT_ISU 872|unnamed3 |
| Organism | Staphylococcus aureus strain ISU 872 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LKVU01000011 (4196..4231 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 4..10, 14..20 (GCGAACG..CGTTCGC) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGCGCCCTT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_ISU 872|unnamed3
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file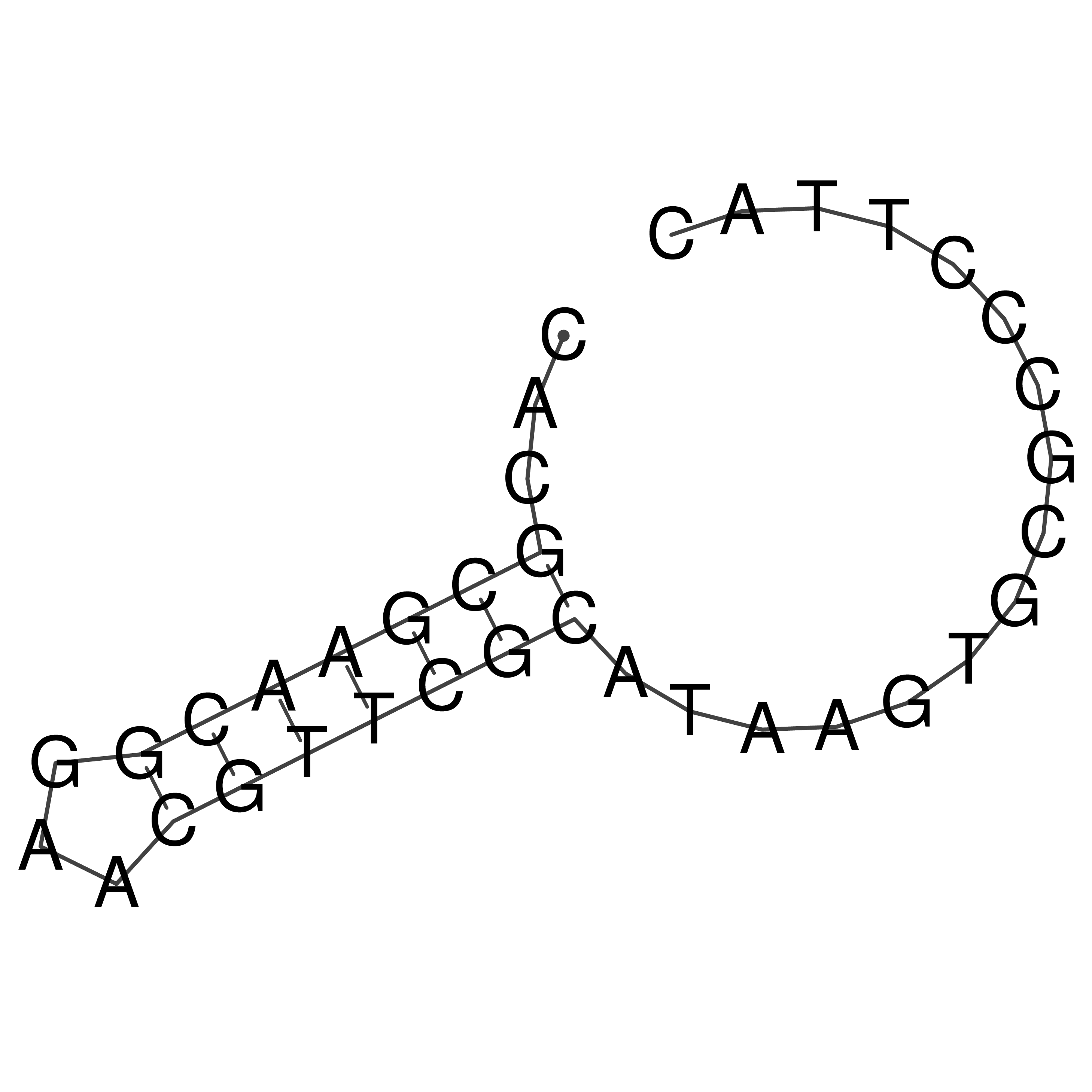
Relaxase
| ID | 1668 | GenBank | WP_223201781 |
| Name | MobA_MobL_APV61_RS01410_ISU 872|unnamed3 |
UniProt ID | _ |
| Length | 663 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 663 a.a. Molecular weight: 78706.59 Da Isoelectric Point: 5.3910
>WP_223201781.1 MULTISPECIES: MobA/MobL family protein [Staphylococcus]
MYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWNKV
HDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRGLD
KNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGWKA
RKFEDETGKKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYVTP
INMYKENERLYDLKQKTSLITDDEDRLNKIEDIEDRQKKLESINEVFEKQAGIFFDKNYPDQSLNYSDDE
KIFITRTILNDRDVLPANNELEDIVKEKRIKEAQISLNTVLGNRDISLESIEKESNFFADKLSNILEKNN
LSFDDVLENKHEGMEDSLKIDYYTNKLEVFRNAENILEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQQL
IDFKTYHGTENTIEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLSEYDF
DYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLINAGLDSMIFNFNEI
FRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
MYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWNKV
HDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRGLD
KNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGWKA
RKFEDETGKKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYVTP
INMYKENERLYDLKQKTSLITDDEDRLNKIEDIEDRQKKLESINEVFEKQAGIFFDKNYPDQSLNYSDDE
KIFITRTILNDRDVLPANNELEDIVKEKRIKEAQISLNTVLGNRDISLESIEKESNFFADKLSNILEKNN
LSFDDVLENKHEGMEDSLKIDYYTNKLEVFRNAENILEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQQL
IDFKTYHGTENTIEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLSEYDF
DYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLINAGLDSMIFNFNEI
FRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 2581 | GenBank | NZ_LKVU01000011 |
| Plasmid name | ISU 872|unnamed3 | Incompatibility group | - |
| Plasmid size | 18958 bp | Coordinate of oriT [Strand] | 4196..4231 [-] |
| Host baterium | Staphylococcus aureus strain ISU 872 |
Cargo genes
| Drug resistance gene | aac(6')-aph(2'') |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |