Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102068 |
| Name | oriT_KP41|unnamed1 KP41_35 |
| Organism | Klebsiella pneumoniae strain KP41 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MOLX01000035 (51931..52036 [+], 106 nt) |
| oriT length | 106 nt |
| IRs (inverted repeats) | 30..35, 41..46 (CCGCCG..CGGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 106 nt
>oriT_KP41|unnamed1 KP41_35
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
AGTACGGGACAAGATGTGTTTTTGGAGTACCGCCGACACGCGGCGGCCGTCCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTATCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file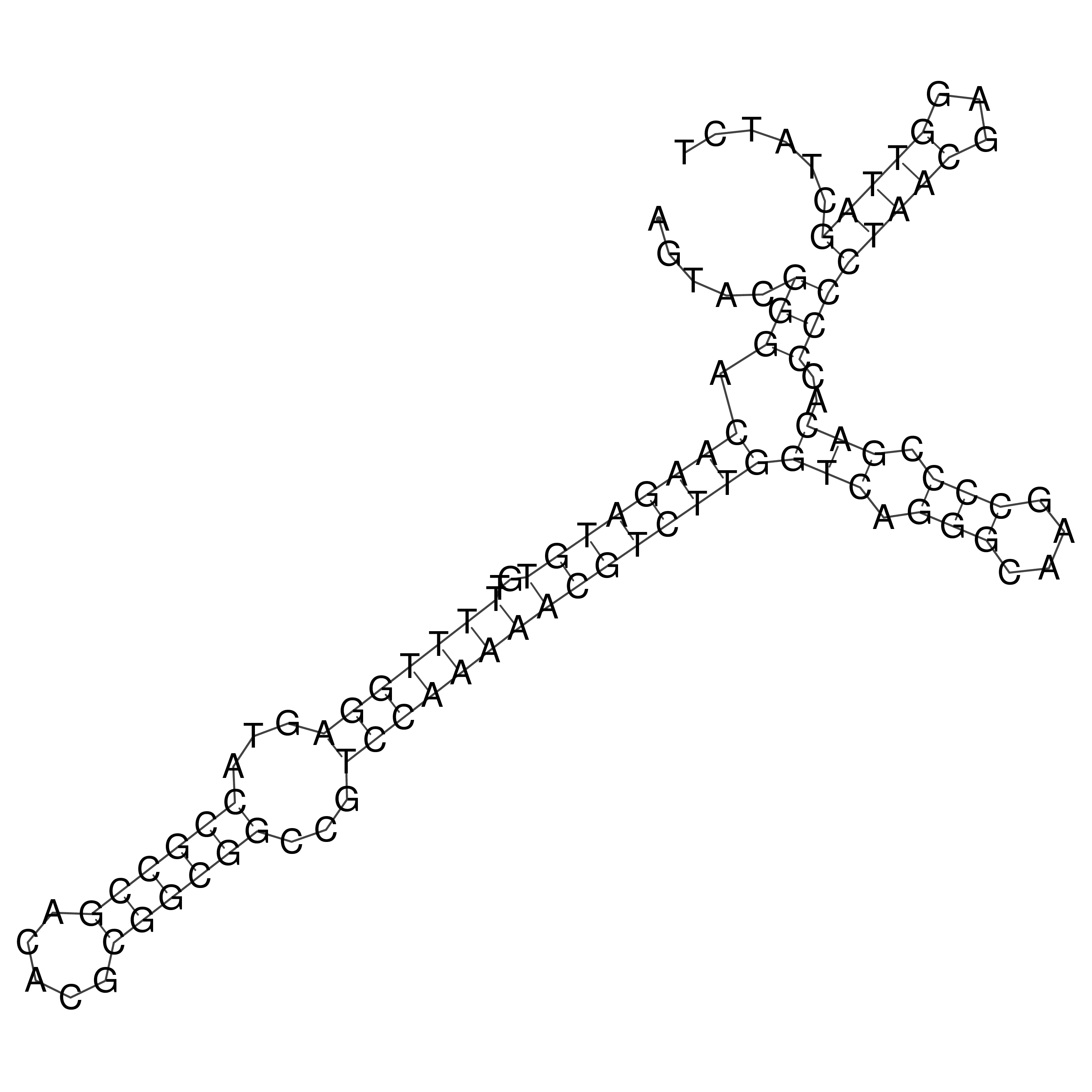
Relaxase
| ID | 1651 | GenBank | WP_004187323 |
| Name | Relaxase_BLJ97_RS24775_KP41|unnamed1 KP41_35 |
UniProt ID | A0A3U8KUL3 |
| Length | 659 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 659 a.a. Molecular weight: 75179.89 Da Isoelectric Point: 9.9948
>WP_004187323.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacterales]
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQFRSESDDKFNPK
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFADIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQFRSESDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3U8KUL3 |
T4CP
| ID | 1279 | GenBank | WP_004187443 |
| Name | t4cp2_BLJ97_RS24515_KP41|unnamed1 KP41_35 |
UniProt ID | _ |
| Length | 695 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 695 a.a. Molecular weight: 79519.87 Da Isoelectric Point: 4.8309
>WP_004187443.1 MULTISPECIES: hypothetical protein [Enterobacterales]
MQQESVDSKKVIRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYKRARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFPEDSGDIRMDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGK
KEEVLLTHALNNQAPYGLIYDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANT
RVKYFESIEDRKTFEILRETVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQG
VISFQDALVRSAAFYIPDNEKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMHFPPIK
LFGYDKAANSLLEKIEVFYELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
MQQESVDSKKVIRPDGSFMEWFLTPNVQFGLLAILTVLGLFLPFTMLLSILILPPLMVAFTDRKFRPPLR
MPKDCEMFDDTLTTETQAEYRLGPIRIPHKVRERKKAKGILYVGYERGRLFGRELWLNMTDLLRHMVFFG
TTGSGKTETFYGFIVNFLLWCRGYCLSDGKADNKLAFATWSLARRFGREDDYYVLNLLTGSIDRFVNLVK
QESIPAQSNSVNLFSVAPPTFIIQLMESMLPQVGGDSAQWQDTAKAMMSALINALCYKRARGELLLSQRT
IQKNMSLPAMAALYVEAKKNGWHSEGYAALESYLENTPGFLLANAEYPETWEARAFEQHNYMSRQFLKTL
SLFNETYGHVFPEDSGDIRMDDIFHNDRILIVMIPSLELSRGEAATLGRLYVTLQRMTISKDLGYQLEGK
KEEVLLTHALNNQAPYGLIYDELGQYFTSGMDTLSAQMRSLEKMGVFSSQDHPSLARGANGEVDSLIANT
RVKYFESIEDRKTFEILRETVGQDYYSELSGQEAEHGTFSKTVREGDLYQIREKDRVNIRELRKQTEGQG
VISFQDALVRSAAFYIPDNEKFSSELPMRINRFIEVLPPSPATLYSIYPERKDDELAETNPDAMHFPPIK
LFGYDKAANSLLEKIEVFYELQCGAISPEEMSVCLFELFAEWLDGTETDDVLYHQEDDLFPMIEM
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1882..23595
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BLJ97_RS24425 (BLJ97_24445) | 1..1916 | + | 1916 | WP_227818823 | LPD7 domain-containing protein | - |
| BLJ97_RS24430 (BLJ97_24450) | 1882..2394 | + | 513 | WP_011091071 | hypothetical protein | traL |
| BLJ97_RS29980 | 2395..2607 | - | 213 | WP_305953721 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| BLJ97_RS24435 (BLJ97_24455) | 2579..2989 | - | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| BLJ97_RS24440 (BLJ97_24460) | 3057..3770 | + | 714 | WP_004187467 | DotI/IcmL family type IV secretion protein | traM |
| BLJ97_RS29985 | 3779..4930 | + | 1152 | WP_004187471 | DotH/IcmK family type IV secretion protein | traN |
| BLJ97_RS24455 (BLJ97_24475) | 4942..6291 | + | 1350 | WP_004187474 | conjugal transfer protein TraO | traO |
| BLJ97_RS24460 (BLJ97_24480) | 6303..7007 | + | 705 | WP_015060002 | conjugal transfer protein TraP | traP |
| BLJ97_RS24465 (BLJ97_24485) | 7031..7561 | + | 531 | WP_004187478 | conjugal transfer protein TraQ | traQ |
| BLJ97_RS24470 (BLJ97_24490) | 7578..7967 | + | 390 | WP_004187479 | DUF6750 family protein | traR |
| BLJ97_RS24475 (BLJ97_24495) | 8013..8507 | + | 495 | WP_004187480 | hypothetical protein | - |
| BLJ97_RS24480 (BLJ97_24500) | 8504..11554 | + | 3051 | WP_004187482 | hypothetical protein | traU |
| BLJ97_RS24485 (BLJ97_24505) | 11551..12759 | + | 1209 | WP_011091082 | conjugal transfer protein TraW | traW |
| BLJ97_RS24490 (BLJ97_24510) | 12756..13352 | + | 597 | WP_015060003 | hypothetical protein | - |
| BLJ97_RS24495 (BLJ97_24515) | 13345..15540 | + | 2196 | WP_015062834 | DotA/TraY family protein | traY |
| BLJ97_RS24500 (BLJ97_24520) | 15542..16195 | + | 654 | WP_044706695 | hypothetical protein | - |
| BLJ97_RS24505 (BLJ97_24525) | 16264..16506 | + | 243 | WP_023893611 | IncL/M type plasmid replication protein RepC | - |
| BLJ97_RS24510 (BLJ97_24530) | 16802..17857 | + | 1056 | WP_015060006 | plasmid replication initiator RepA | - |
| BLJ97_RS32250 | 19081..19176 | + | 96 | WP_004206884 | DinQ-like type I toxin DqlB | - |
| BLJ97_RS24515 (BLJ97_24535) | 19227..21314 | - | 2088 | WP_004187443 | hypothetical protein | - |
| BLJ97_RS24520 (BLJ97_24540) | 21327..22277 | - | 951 | WP_004187441 | DsbC family protein | trbB |
| BLJ97_RS24525 (BLJ97_24545) | 22288..23595 | - | 1308 | WP_015062796 | hypothetical protein | trbA |
| BLJ97_RS24530 (BLJ97_24550) | 23595..23990 | - | 396 | WP_004187436 | lytic transglycosylase domain-containing protein | - |
| BLJ97_RS24535 (BLJ97_24555) | 24095..24478 | - | 384 | WP_032451295 | DUF1496 domain-containing protein | - |
| BLJ97_RS24540 (BLJ97_24560) | 24558..25211 | + | 654 | WP_039592068 | type II CAAX endopeptidase family protein | - |
| BLJ97_RS24545 (BLJ97_24565) | 25364..25798 | + | 435 | WP_015062799 | translesion error-prone DNA polymerase V autoproteolytic subunit | - |
| BLJ97_RS24550 (BLJ97_24570) | 25786..27051 | + | 1266 | WP_040224115 | translesion error-prone DNA polymerase V subunit UmuC | - |
| BLJ97_RS24555 (BLJ97_24575) | 27074..27922 | - | 849 | WP_021561452 | 3'-5' exonuclease | - |
| BLJ97_RS24560 (BLJ97_24580) | 27925..28245 | - | 321 | WP_021561451 | hypothetical protein | - |
Host bacterium
| ID | 2512 | GenBank | NZ_MOLX01000035 |
| Plasmid name | KP41|unnamed1 KP41_35 | Incompatibility group | IncL/M |
| Plasmid size | 58558 bp | Coordinate of oriT [Strand] | 51931..52036 [+] |
| Host baterium | Klebsiella pneumoniae strain KP41 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA8 |