Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102051 |
| Name | oriT_UCI 7|unnamed1 |
| Organism | Staphylococcus aureus strain UCI 7 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LKYY01000101 (2370..2407 [+], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 14..22, 29..37 (TAAAGTATA..TATACTTTA) 2..8, 13..19 (ACTTTAT..ATAAAGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_UCI 7|unnamed1
CACTTTATGAATATAAAGTATAGTCTGTTATACTTTAC
CACTTTATGAATATAAAGTATAGTCTGTTATACTTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file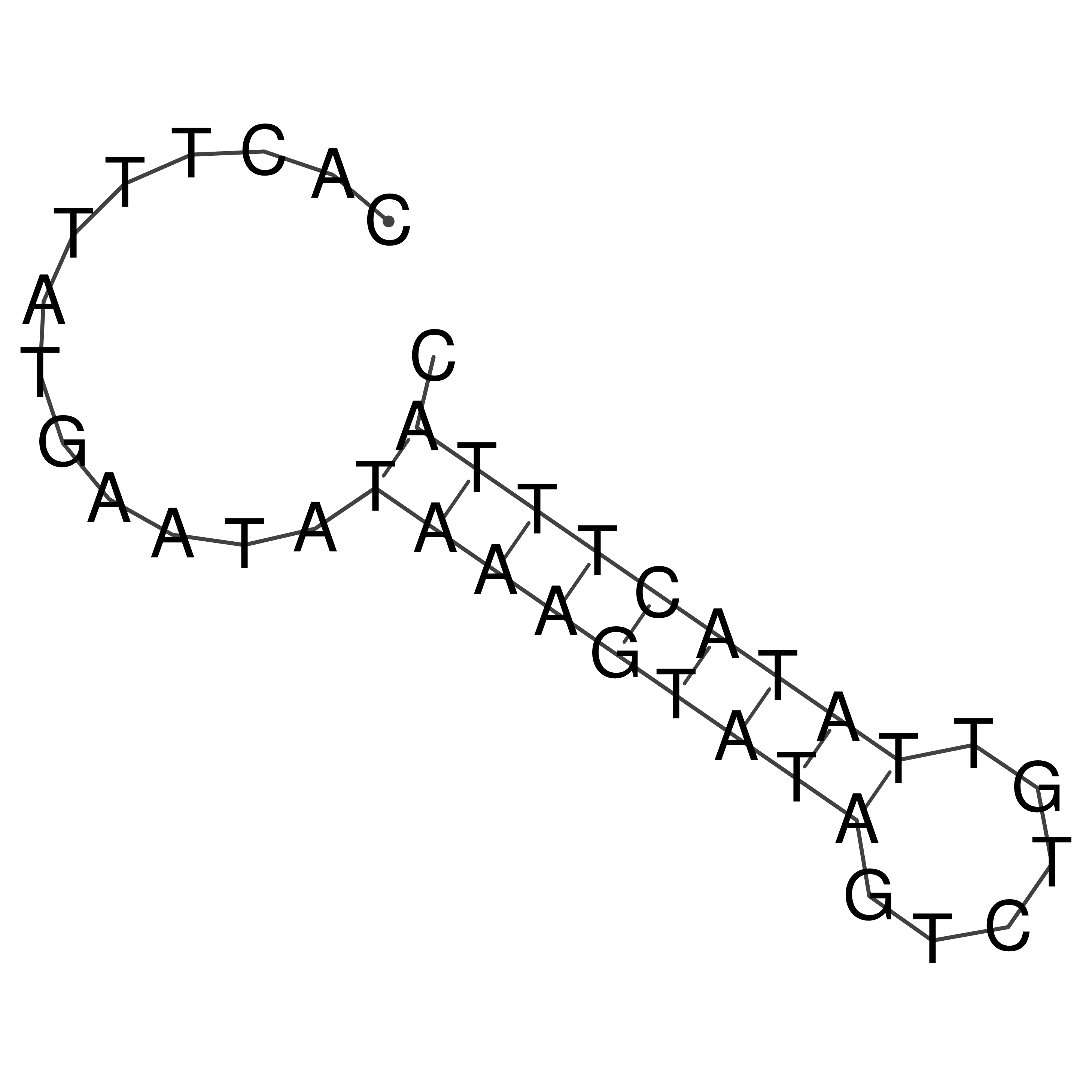
Relaxase
| ID | 1638 | GenBank | WP_000119405 |
| Name | mobV_APW43_RS10360_UCI 7|unnamed1 |
UniProt ID | F5X340 |
| Length | 420 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 420 a.a. Molecular weight: 49660.33 Da Isoelectric Point: 9.9693
>WP_000119405.1 MULTISPECIES: MobV family relaxase [Bacteria]
MSYAVCRMQKVKSAGLKGMQFHNQRERKSRTNDDIDHERTRENYDLKNDKNIDYNERVKEIIESQKTGTR
KTRKDAVLVNELLVTSDRDFFEQLDPGEQKRFFEESYKLFSERYGKQNIAYATVHNDEQTPHMHLGVVPM
RDGKLQGKNVFNRQELLWLQDKFPEHMKKQGFELKRGERGSDRKHIETAKFKKQTLEKEIDFLEKNLAVK
KDEWTAYSDKVKSDLEVPAKRHMKSVEVPTGEKSMFGLGKEIMKTEKKPTKNVVISERDYKNLVTAARDN
DRLKQHVRNLMSTDMAREYKKLSKEHGQVKEKYSGLVERFNENVNDYNELLEENKSLKSKISDLKRDVSL
IYESTKEFLKERTDGLKAFKNVFKGFVDKVKDKTAQFQEKHDLEPKKNEFELTHNREVKKERSRDQGMSL
MSYAVCRMQKVKSAGLKGMQFHNQRERKSRTNDDIDHERTRENYDLKNDKNIDYNERVKEIIESQKTGTR
KTRKDAVLVNELLVTSDRDFFEQLDPGEQKRFFEESYKLFSERYGKQNIAYATVHNDEQTPHMHLGVVPM
RDGKLQGKNVFNRQELLWLQDKFPEHMKKQGFELKRGERGSDRKHIETAKFKKQTLEKEIDFLEKNLAVK
KDEWTAYSDKVKSDLEVPAKRHMKSVEVPTGEKSMFGLGKEIMKTEKKPTKNVVISERDYKNLVTAARDN
DRLKQHVRNLMSTDMAREYKKLSKEHGQVKEKYSGLVERFNENVNDYNELLEENKSLKSKISDLKRDVSL
IYESTKEFLKERTDGLKAFKNVFKGFVDKVKDKTAQFQEKHDLEPKKNEFELTHNREVKKERSRDQGMSL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | F5X340 |
Host bacterium
| ID | 2495 | GenBank | NZ_LKYY01000101 |
| Plasmid name | UCI 7|unnamed1 | Incompatibility group | - |
| Plasmid size | 5432 bp | Coordinate of oriT [Strand] | 2370..2407 [+] |
| Host baterium | Staphylococcus aureus strain UCI 7 |
Cargo genes
| Drug resistance gene | aadD |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |