Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 102009 |
| Name | oriT_ISU 918|unnamed1 |
| Organism | Staphylococcus aureus strain ISU 918 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LKWR01000043 (2860..3063 [+], 204 nt) |
| oriT length | 204 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 204 nt
>oriT_ISU 918|unnamed1
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file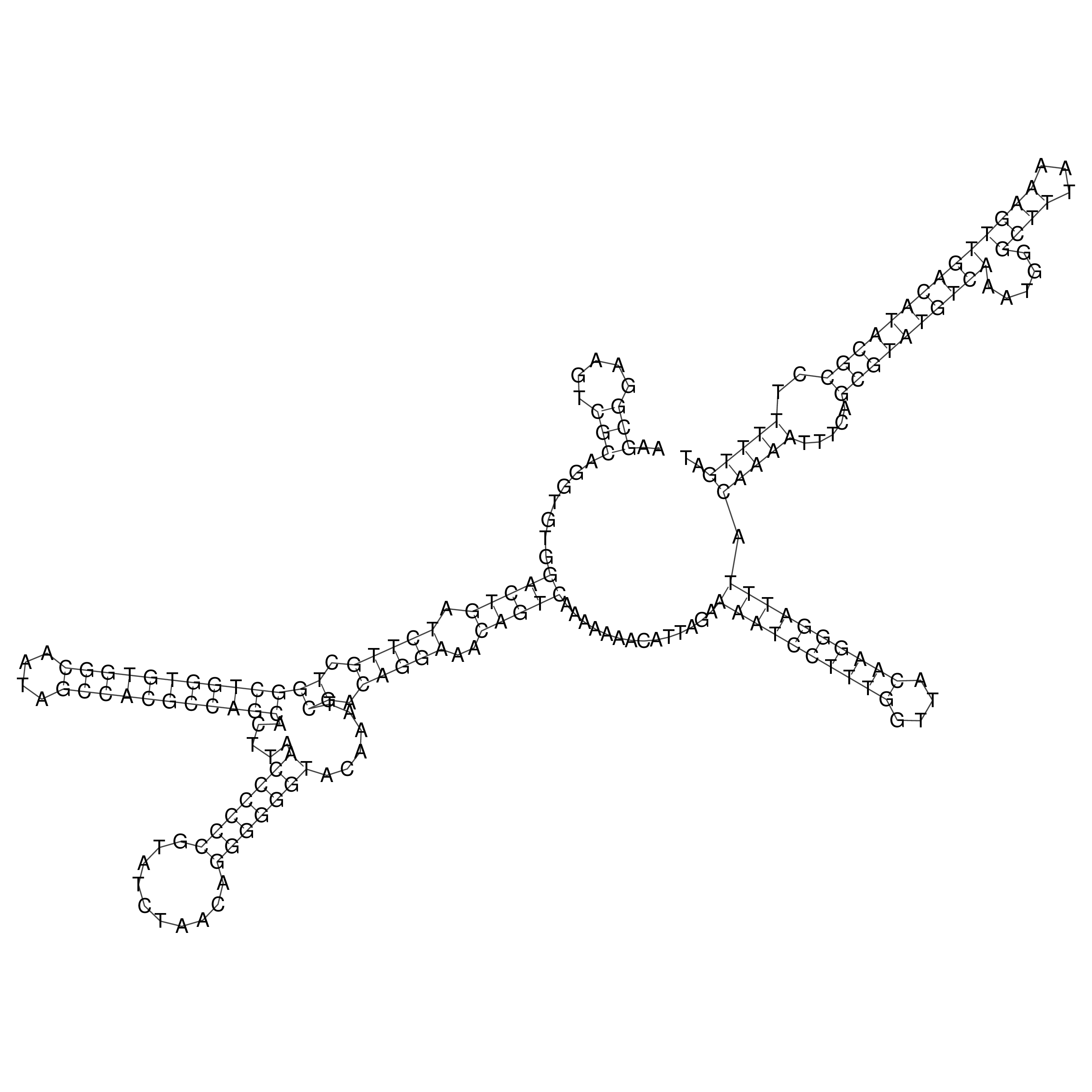
Relaxase
| ID | 1603 | GenBank | WP_031919544 |
| Name | mobT_APV79_RS07030_ISU 918|unnamed1 |
UniProt ID | _ |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47432.12 Da Isoelectric Point: 6.5091
>WP_031919544.1 MobT family relaxase [Staphylococcus aureus]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDFQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDFQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1264 | GenBank | WP_000813488 |
| Name | tcpA_APV79_RS07020_ISU 918|unnamed1 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>WP_000813488.1 MULTISPECIES: FtsK/SpoIIIE domain-containing protein [Bacteria]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 755..12165
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| APV79_RS07000 (APV79_08060) | 1..386 | + | 386 | WP_142990993 | transposase | - |
| APV79_RS07010 (APV79_08065) | 755..1069 | + | 315 | WP_000420682 | YdcP family protein | orf23 |
| APV79_RS07015 (APV79_08070) | 1085..1471 | + | 387 | WP_000985015 | YdcP family protein | orf23 |
| APV79_RS07020 (APV79_08075) | 1500..2885 | + | 1386 | WP_000813488 | FtsK/SpoIIIE domain-containing protein | virb4 |
| APV79_RS07025 (APV79_08080) | 2888..3040 | + | 153 | WP_000879507 | hypothetical protein | - |
| APV79_RS07030 (APV79_08085) | 3063..4268 | + | 1206 | WP_031919544 | MobT family relaxase | - |
| APV79_RS07035 (APV79_08090) | 4311..4532 | + | 222 | WP_001009056 | hypothetical protein | orf19 |
| APV79_RS07040 (APV79_08095) | 4649..5146 | + | 498 | WP_000342539 | antirestriction protein ArdA | - |
| APV79_RS07045 (APV79_08100) | 5235..5627 | + | 393 | WP_000723888 | conjugal transfer protein | orf17a |
| APV79_RS07050 (APV79_08105) | 5611..8058 | + | 2448 | WP_071938142 | ATP-binding protein | virb4 |
| APV79_RS07055 (APV79_08110) | 8061..10238 | + | 2178 | WP_000804748 | membrane protein | orf15 |
| APV79_RS07060 (APV79_08115) | 10235..11236 | + | 1002 | WP_000769868 | bifunctional lysozyme/C40 family peptidase | orf14 |
| APV79_RS07065 (APV79_08120) | 11233..12165 | + | 933 | WP_001224318 | conjugal transfer protein | orf13 |
| APV79_RS07070 | 12410..12526 | + | 117 | WP_001814923 | tetracycline resistance determinant leader peptide | - |
| APV79_RS07075 (APV79_08125) | 12542..14461 | + | 1920 | WP_000691736 | tetracycline resistance ribosomal protection protein Tet(M) | - |
| APV79_RS07080 | 14580..14747 | + | 168 | WP_000336323 | cysteine-rich KTR domain-containing protein | - |
| APV79_RS07085 (APV79_08130) | 14807..15160 | - | 354 | WP_001227347 | helix-turn-helix transcriptional regulator | - |
Host bacterium
| ID | 2453 | GenBank | NZ_LKWR01000043 |
| Plasmid name | ISU 918|unnamed1 | Incompatibility group | - |
| Plasmid size | 15266 bp | Coordinate of oriT [Strand] | 2860..3063 [+] |
| Host baterium | Staphylococcus aureus strain ISU 918 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |