Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101993 |
| Name | oriT_pADP_1 |
| Organism | Pseudomonas sp. EGD-AKN5 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_LZYC01000023 (39618..39716 [-], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_pADP_1
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file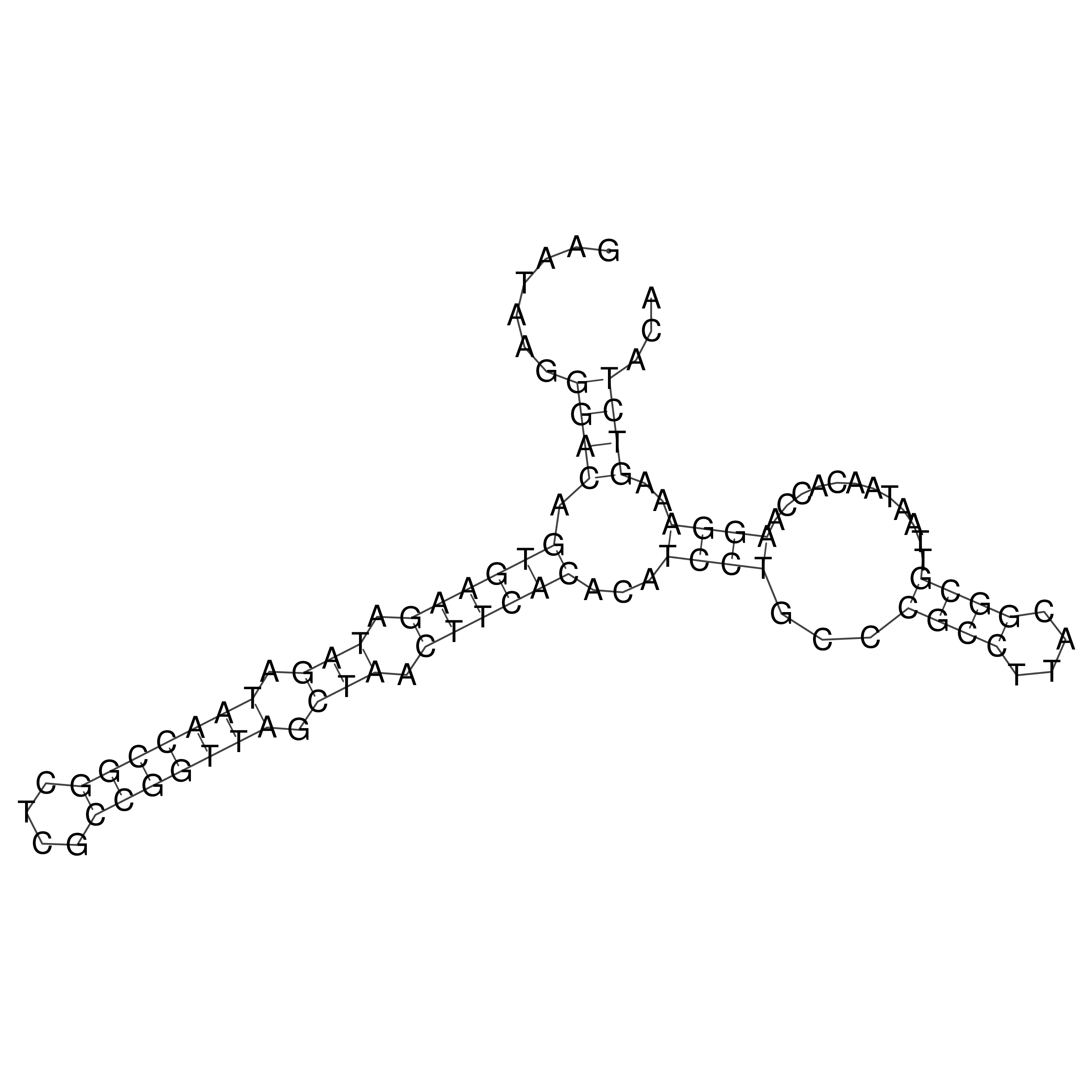
Relaxase
| ID | 1592 | GenBank | WP_011117146 |
| Name | Relaxase_BAE52_RS04910_pADP_1 |
UniProt ID | Q937B9 |
| Length | 746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 746 a.a. Molecular weight: 82120.82 Da Isoelectric Point: 10.8213
>WP_011117146.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Pseudomonadota]
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWVKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQNLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
IQAINKQFQQERTALYAEHRRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWVKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQNLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
IQAINKQFQQERTALYAEHRRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q937B9 |
T4CP
| ID | 1254 | GenBank | WP_011114047 |
| Name | t4cp2_BAE52_RS04715_pADP_1 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93980.15 Da Isoelectric Point: 6.3901
>WP_011114047.1 MULTISPECIES: VirB4 family type IV secretion/conjugal transfer ATPase [Bacteria]
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1255 | GenBank | WP_000694765 |
| Name | t4cp2_BAE52_RS04915_pADP_1 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69897.09 Da Isoelectric Point: 8.2993
>WP_000694765.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Bacteria]
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 917..6926
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BAE52_RS04695 (BAE52_00015) | 1..421 | - | 421 | Protein_0 | conjugal transfer protein TrbI | - |
| BAE52_RS04700 (BAE52_00020) | 426..914 | - | 489 | WP_006122479 | conjugal transfer protein TrbH | - |
| BAE52_RS04705 (BAE52_00025) | 917..1816 | - | 900 | WP_006122480 | P-type conjugative transfer protein TrbG | virB9 |
| BAE52_RS04710 (BAE52_00030) | 1834..2616 | - | 783 | WP_011114048 | conjugal transfer protein TrbF | virB8 |
| BAE52_RS04715 (BAE52_00035) | 2613..5171 | - | 2559 | WP_011114047 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| BAE52_RS04720 (BAE52_00040) | 5168..5479 | - | 312 | WP_001207961 | conjugal transfer protein TrbD | virB3 |
| BAE52_RS04725 (BAE52_00045) | 5483..5947 | - | 465 | WP_011114046 | conjugal transfer system pilin TrbC | virB2 |
| BAE52_RS04730 (BAE52_00050) | 5964..6926 | - | 963 | WP_000132020 | P-type conjugative transfer ATPase TrbB | virB11 |
| BAE52_RS04735 (BAE52_00055) | 7236..7598 | - | 363 | WP_011114045 | transcriptional regulator | - |
| BAE52_RS04740 (BAE52_00060) | 7712..8053 | + | 342 | WP_000019167 | single-stranded DNA-binding protein | - |
| BAE52_RS04745 (BAE52_00065) | 8100..9320 | + | 1221 | WP_011114072 | plasmid replication initiator TrfA | - |
| BAE52_RS04750 (BAE52_00070) | 9716..11086 | - | 1371 | WP_011117183 | IS1182-like element ISPsp5 family transposase | - |
Host bacterium
| ID | 2437 | GenBank | NZ_LZYC01000023 |
| Plasmid name | pADP_1 | Incompatibility group | IncP1 |
| Plasmid size | 63629 bp | Coordinate of oriT [Strand] | 39618..39716 [-] |
| Host baterium | Pseudomonas sp. EGD-AKN5 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |