Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101900 |
| Name | oriT_FWSEC0423|unnamed10 |
| Organism | Escherichia coli strain FWSEC0423 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_RROS01000138 (9526..9598 [+], 73 nt) |
| oriT length | 73 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 73 nt
>oriT_FWSEC0423|unnamed10
GTCGGGGCAAAGCCCTGACCAGGTGGCATTTGTCTGATTGCGCGTGCGCGGTCCGACAAATGCACATCCTGTC
GTCGGGGCAAAGCCCTGACCAGGTGGCATTTGTCTGATTGCGCGTGCGCGGTCCGACAAATGCACATCCTGTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file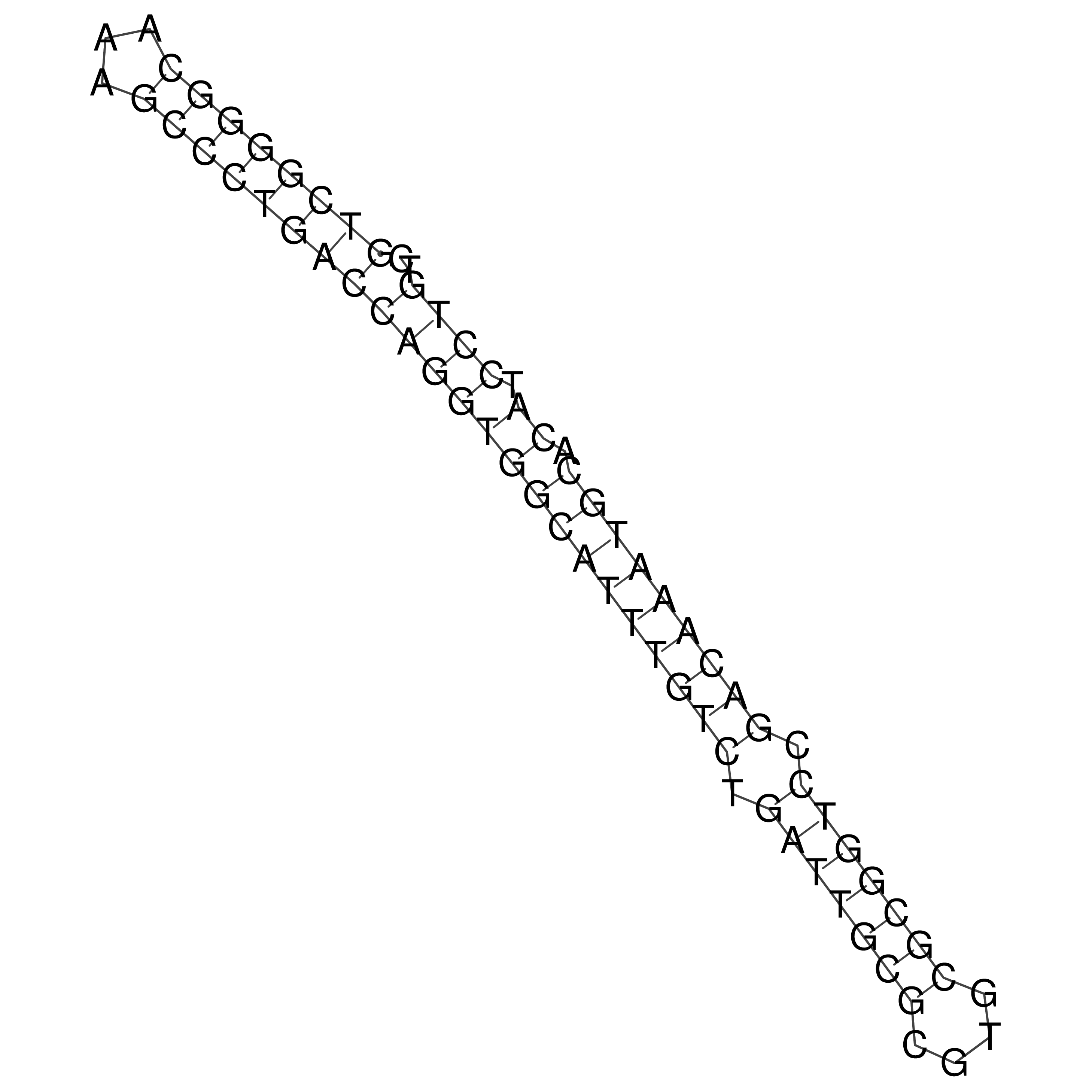
Relaxase
| ID | 1553 | GenBank | WP_062891657 |
| Name | Relaxase_C9198_RS27585_FWSEC0423|unnamed10 |
UniProt ID | _ |
| Length | 906 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 105092.43 Da Isoelectric Point: 8.3694
>WP_062891657.1 relaxase/mobilization nuclease domain-containing protein [Escherichia coli]
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWINFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPVSGHINRLSWSQERLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPKMARNGELKSCLDVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGHSIGTQPVNDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPELRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAHIRVQFRDPQVRKLHYHIAEVQRMQALIRLKENVREERLALMAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHNDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTISRPDVDLHRQRSEEYYHEYIRQQTRLSESHDENYSSRQENTWVPPSPGM
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWINFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPVSGHINRLSWSQERLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPKMARNGELKSCLDVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWQPVPKDIFTQVIPGERFRGHSIGTQPVNDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPELRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAHIRVQFRDPQVRKLHYHIAEVQRMQALIRLKENVREERLALMAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHNDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTISRPDVDLHRQRSEEYYHEYIRQQTRLSESHDENYSSRQENTWVPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 628 | GenBank | WP_001291058 |
| Name | WP_001291058_FWSEC0423|unnamed10 |
UniProt ID | A0A7I0L241 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12730.52 Da Isoelectric Point: 9.9656
>WP_001291058.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0L241 |
T4CP
| ID | 1227 | GenBank | WP_169072845 |
| Name | t4cp2_C9198_RS27570_FWSEC0423|unnamed10 |
UniProt ID | _ |
| Length | 422 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 422 a.a. Molecular weight: 47602.68 Da Isoelectric Point: 5.5539
>WP_169072845.1 TraM recognition domain-containing protein, partial [Escherichia coli]
SGQFSETFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMILARD
LGYRLEGTDAQTLEVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMMAQDQERIEGQTSAT
NTATLMQNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQ
DLIRLNAGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPE
HVSSIIGVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNT
TREERRVCYITLNRPEEEYSGPEDIPSVKEILNTLLPLEILLPVAEKNKSQTINNGHPQKSYGAQPTTDE
KY
SGQFSETFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFVTQQSMILARD
LGYRLEGTDAQTLEVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMMAQDQERIEGQTSAT
NTATLMQNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQ
DLIRLNAGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYISVEAPRLERLRRLVPRTVQRRLPTPE
HVSSIIGVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNT
TREERRVCYITLNRPEEEYSGPEDIPSVKEILNTLLPLEILLPVAEKNKSQTINNGHPQKSYGAQPTTDE
KY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 2344 | GenBank | NZ_RROS01000138 |
| Plasmid name | FWSEC0423|unnamed10 | Incompatibility group | - |
| Plasmid size | 10028 bp | Coordinate of oriT [Strand] | 9526..9598 [+] |
| Host baterium | Escherichia coli strain FWSEC0423 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |