Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101855 |
| Name | oriT_pDCCK-1.1 |
| Organism | Enterobacter hormaechei strain HA16Eho |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JAMQJW010000022 (44378..44482 [-], 105 nt) |
| oriT length | 105 nt |
| IRs (inverted repeats) | 32..37, 40..45 (GCCGGC..GCCGGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 105 nt
>oriT_pDCCK-1.1
AGTACGGGACAAGATGTGTTTTTGGTACACCGCCGGCACGCCGGCAGTGACGCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTAT
AGTACGGGACAAGATGTGTTTTTGGTACACCGCCGGCACGCCGGCAGTGACGCAAAAACGTCTTGGTCAGGGCAAGCCCCGACACCCCCTAACGAGGTTAGCTAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file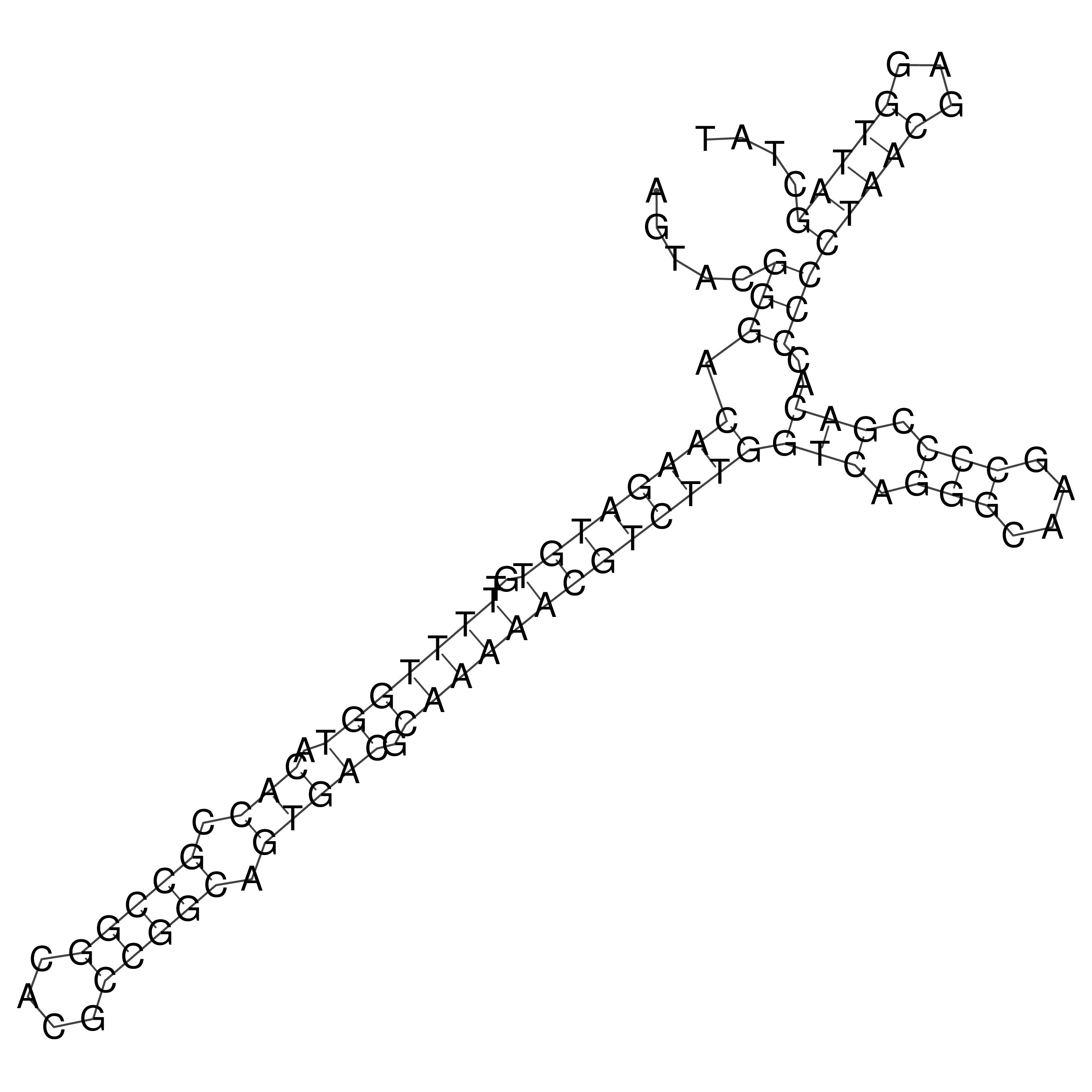
Relaxase
| ID | 1533 | GenBank | WP_004206924 |
| Name | Relaxase_NDQ76_RS20130_pDCCK-1.1 |
UniProt ID | A0A2V4FJ40 |
| Length | 659 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 659 a.a. Molecular weight: 75087.84 Da Isoelectric Point: 10.0285
>WP_004206924.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacterales]
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFAGIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQLRSESDDKFNPK
MVIPKIIEGRRDKKSSFGQLIKYMADKPSQELTDTVQPTPETALAVKSDMFEGLNNYLTRKQKVISTPVD
VEPGVQRVVVGDVTCQYNTFSLDGAAQEMNSVSQQSTRCKDPVMHYVLSWPDYEKPNDDQVFDSVKFTLA
SMGMSDHQYVAAIHRDTDNLHVHVAVNRINPQTYKAASSSFTKDTLHQACRLLELKNGWSHSNGAYVVND
RQQIVRNPHSKKERGNWRSLDRINKMENKEGVETLYRYIVGDEQVGGSRQNLIHVSAGLREAKSWDDVHK
TFAGIGLRVEKAQGKKGYVITHEHQNQKTAVKASLVFNKAQYTLKSMEERFGEYQPSHIEPAKVSVFKTA
YTPGAYRRDANKRLQRKIERAEERMLLKGRYRAYRNNLPIYSPDKDRIADEYRKIAQHTRLVKNNVRHSV
SDPHTRKLMYNLAEFKRLQAVANLRLSLREERNGFRAANPRLSYREWVEQEALKGDKAALSQMRGFAYSS
RKKEKYKQQLVEQIGFNRTFNAITSHDRDDVAVMASARHGVKPRLLKDGTVIFERDGKPVAADRGHIVLT
ESNGIDKEKTADLAIALTIAGKAKSVRVDGDGEFKELCCNRIVDAAVNHNHPVAQGITFTDAAQQAYAQN
EKHRLIREQNNSKNEMQLRSESDDKFNPK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A2V4FJ40 |
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 22384..41580
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NDQ76_RS20000 (NDQ76_20000) | 17796..17984 | - | 189 | Protein_17 | MerC family mercury resistance protein | - |
| NDQ76_RS20005 (NDQ76_20005) | 18012..18287 | - | 276 | WP_000732275 | mercury resistance system periplasmic binding protein MerP | - |
| NDQ76_RS20010 (NDQ76_20010) | 18303..18653 | - | 351 | WP_031623830 | mercuric ion transporter MerT | - |
| NDQ76_RS20015 (NDQ76_20015) | 18725..19180 | + | 456 | WP_001166628 | Hg(II)-responsive transcriptional regulator | - |
| NDQ76_RS20020 (NDQ76_20020) | 20072..21127 | - | 1056 | WP_230206039 | plasmid replication initiator RepA | - |
| NDQ76_RS20025 (NDQ76_20025) | 21424..21654 | - | 231 | WP_004187496 | IncL/M type plasmid replication protein RepC | - |
| NDQ76_RS20030 (NDQ76_20030) | 21728..22381 | - | 654 | WP_004206935 | hypothetical protein | - |
| NDQ76_RS20035 (NDQ76_20035) | 22384..24564 | - | 2181 | WP_004187492 | DotA/TraY family protein | traY |
| NDQ76_RS20040 (NDQ76_20040) | 24557..25207 | - | 651 | WP_004187488 | hypothetical protein | - |
| NDQ76_RS20045 (NDQ76_20045) | 25204..26412 | - | 1209 | WP_004187486 | conjugal transfer protein TraW | traW |
| NDQ76_RS20050 (NDQ76_20050) | 26409..29459 | - | 3051 | WP_011154474 | conjugative transfer protein | traU |
| NDQ76_RS20055 (NDQ76_20055) | 29456..29950 | - | 495 | WP_004187480 | hypothetical protein | - |
| NDQ76_RS20060 (NDQ76_20060) | 29996..30385 | - | 390 | WP_011154472 | DUF6750 family protein | traR |
| NDQ76_RS20065 (NDQ76_20065) | 30402..30932 | - | 531 | WP_004187478 | conjugal transfer protein TraQ | traQ |
| NDQ76_RS20070 (NDQ76_20070) | 30956..31660 | - | 705 | WP_004206933 | conjugal transfer protein TraP | traP |
| NDQ76_RS20075 (NDQ76_20075) | 31672..33021 | - | 1350 | WP_004206932 | conjugal transfer protein TraO | traO |
| NDQ76_RS20080 (NDQ76_20080) | 33033..34184 | - | 1152 | WP_015062843 | DotH/IcmK family type IV secretion protein | traN |
| NDQ76_RS20085 (NDQ76_20085) | 34193..34906 | - | 714 | WP_223294958 | DotI/IcmL family type IV secretion protein | traM |
| NDQ76_RS20090 (NDQ76_20090) | 34974..35384 | + | 411 | WP_004187465 | H-NS family nucleoid-associated regulatory protein | - |
| NDQ76_RS20095 (NDQ76_20095) | 35413..35568 | + | 156 | WP_172693602 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| NDQ76_RS20100 (NDQ76_20100) | 35569..36081 | - | 513 | WP_011091071 | hypothetical protein | traL |
| NDQ76_RS20105 (NDQ76_20105) | 36047..39352 | - | 3306 | WP_242781107 | LPD7 domain-containing protein | - |
| NDQ76_RS20110 (NDQ76_20110) | 39377..39637 | - | 261 | WP_004187310 | IcmT/TraK family protein | traK |
| NDQ76_RS20115 (NDQ76_20115) | 39627..40790 | - | 1164 | WP_004206926 | plasmid transfer ATPase TraJ | virB11 |
| NDQ76_RS20120 (NDQ76_20120) | 40801..41580 | - | 780 | WP_004187315 | type IV secretory system conjugative DNA transfer family protein | traI |
| NDQ76_RS20125 (NDQ76_20125) | 41577..42077 | - | 501 | WP_004206925 | DotD/TraH family lipoprotein | - |
| NDQ76_RS20130 (NDQ76_20130) | 42091..44070 | - | 1980 | WP_004206924 | TraI/MobA(P) family conjugative relaxase | - |
| NDQ76_RS20135 (NDQ76_20135) | 44057..44374 | - | 318 | WP_004206923 | plasmid mobilization protein MobA | - |
| NDQ76_RS20140 (NDQ76_20140) | 44650..45015 | + | 366 | WP_058672219 | hypothetical protein | - |
| NDQ76_RS20145 (NDQ76_20145) | 45516..46052 | - | 537 | WP_004206920 | hypothetical protein | - |
| NDQ76_RS20150 (NDQ76_20150) | 46185..46496 | - | 312 | WP_004187333 | hypothetical protein | - |
Host bacterium
| ID | 2299 | GenBank | NZ_JAMQJW010000022 |
| Plasmid name | pDCCK-1.1 | Incompatibility group | IncL/M |
| Plasmid size | 67953 bp | Coordinate of oriT [Strand] | 44378..44482 [-] |
| Host baterium | Enterobacter hormaechei strain HA16Eho |
Cargo genes
| Drug resistance gene | blaKPC-2 |
| Virulence gene | - |
| Metal resistance gene | merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |