Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101797 |
| Name | oriT_PECIMP|punnamed2 |
| Organism | Enterobacter hormaechei strain PECIMP |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_QHMI01000031 (33573..33663 [+], 91 nt) |
| oriT length | 91 nt |
| IRs (inverted repeats) | 35..42, 45..52 (AGTGCGAT..ATCGCACT) 21..26, 40..45 (TAAATC..GATTTA) |
| Location of nic site | 63..64 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 91 nt
>oriT_PECIMP|punnamed2
TATTTCATTTTTTTGTTTTTTAAATCAGTTAGTTAGTGCGATTTATCGCACTGCGTTAGGTGTATAGCAGGTTAAGGGCTAAAAAATAATC
TATTTCATTTTTTTGTTTTTTAAATCAGTTAGTTAGTGCGATTTATCGCACTGCGTTAGGTGTATAGCAGGTTAAGGGCTAAAAAATAATC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file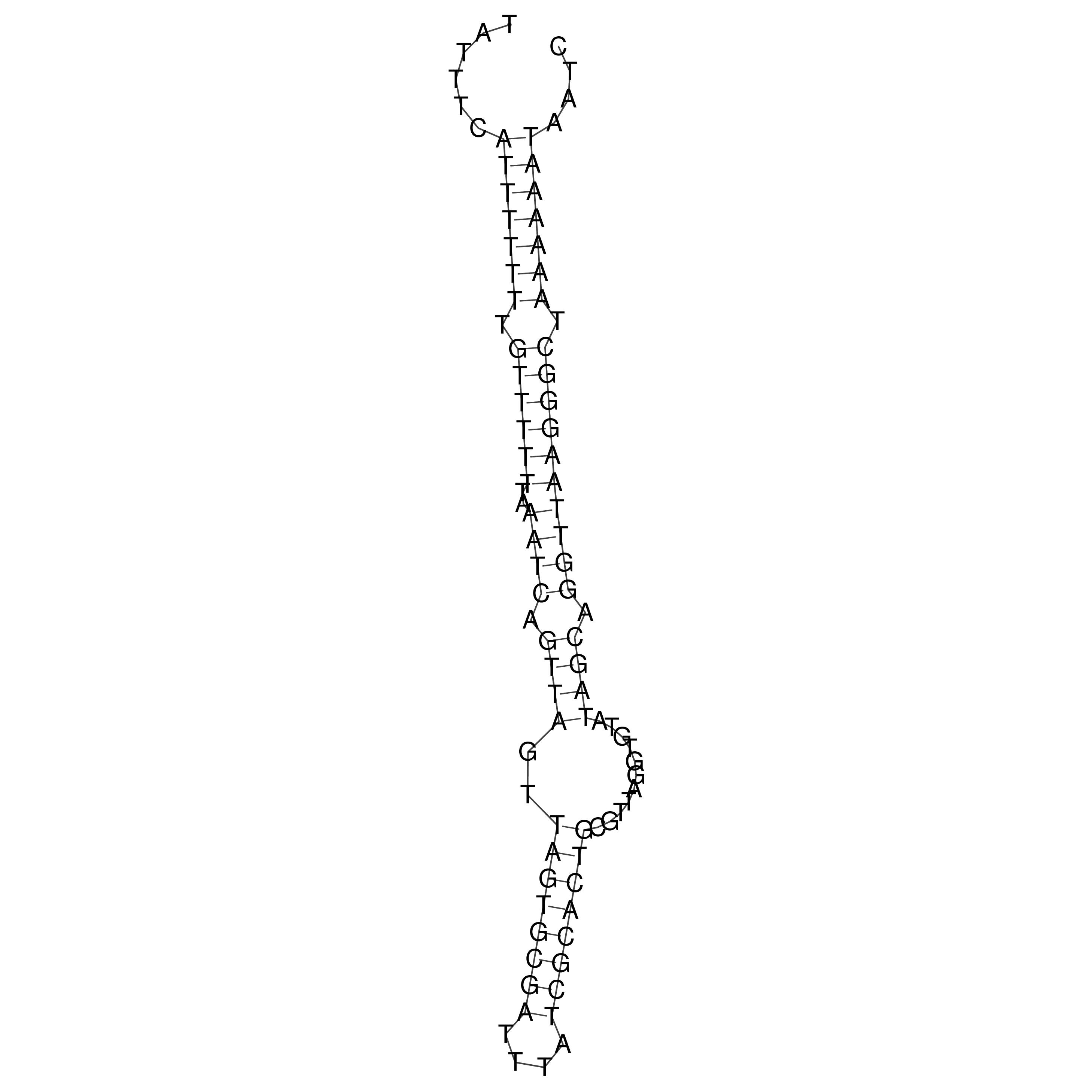
Relaxase
| ID | 1505 | GenBank | WP_058672983 |
| Name | mobF_DL189_RS22760_PECIMP|punnamed2 |
UniProt ID | A0A9X7L0H4 |
| Length | 1072 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1072 a.a. Molecular weight: 118901.56 Da Isoelectric Point: 6.4635
>WP_058672983.1 MULTISPECIES: MobF family relaxase [Enterobacteriaceae]
MLDITTITRIDIASVVGYYADAKDDYYSKDSSFTAWHGQGAEALGLSGEVSSQRFKELLVGDIDPFTQMK
RSSGDATKERLGYDLTFSAPKGVSMQALIHGDKAIIAAHEKAVAAAVQEAEKLAQARTTHAGKTITQNTG
NLVVASFRHETSRALDPELHTHAFVMNMTQRGDGQWRALKNDELMRAKMHLGDVYKQELALELTKAGYEL
RYNNKNNTFDMAHFTDEQIRGFSRRSAQIEEGLAAMGLTRETADAQTKSRVSMATRDKKTDDYSREDIHK
EWVNRANELGIDFNDLSWAGKGKDGPQTSMNTVPNFTSPEVKADRAIQFALKSLSERDASFDQSRLLAVA
NKKAIGQASKQDVEAAYYRAVAKGAIIEGEARYTTTTDQRSKDAPPLPAMTKAEWQAQLTDKKGMNDSQA
KALVAEGIKSGRLKKTSHRVTTVEGIRVEKAVLNIEARGRGKVIAALTSEQVTVQLVGKTLKAEQQKSVV
DICTTTDRFVAAHGFAGTGKSYMTMSAKAVLESQGYNVTALAPYGTQKKSLEDEGMPARTVAAFVKAKDK
KIDEKSVVFIDEAGVIPARQMKVLMETIEKAGARAVFLGDTSQTKAVEAGKPFEQLISAGMQTSYMKDIQ
RQKNDVLLEAVKLAAEGHISGSLARLSNISAEKDQDKRLEAVANRWLSFSPEQREGTLIISGTNESRVIL
NSQIREALQLQGKGVEVNFLERVDSTQAERRDSKYYQVGQIIIPEKDYKNGLQRGESYRVLDTGPGNLLT
VAGSDGQKISFSPRTHKNLSVYTSVAAELAVGDKVMVTRNDKTLDVANGDRFTVTGTTEKGTFTLTNSKG
REIELDQKQAAYLSYAYATTVHKAQGLTCDRVLFNIDTRSLTTSKDVFYVGISRARHEVEIFTDDKKRLL
SSASRNSPKTTASEIDRFLGMEERYKDVNRSYGHSDGQQEKSYGHDAGHQTEEKENTMKNATEAAHDGPD
TSGTENTSQDLRQREQEEVNAAIRYEQRQEATPEMEANPYHDYERYADEGPDYGDMQDYSTYEEYERARD
VELPQHEHENHQHDHDKEGYGL
MLDITTITRIDIASVVGYYADAKDDYYSKDSSFTAWHGQGAEALGLSGEVSSQRFKELLVGDIDPFTQMK
RSSGDATKERLGYDLTFSAPKGVSMQALIHGDKAIIAAHEKAVAAAVQEAEKLAQARTTHAGKTITQNTG
NLVVASFRHETSRALDPELHTHAFVMNMTQRGDGQWRALKNDELMRAKMHLGDVYKQELALELTKAGYEL
RYNNKNNTFDMAHFTDEQIRGFSRRSAQIEEGLAAMGLTRETADAQTKSRVSMATRDKKTDDYSREDIHK
EWVNRANELGIDFNDLSWAGKGKDGPQTSMNTVPNFTSPEVKADRAIQFALKSLSERDASFDQSRLLAVA
NKKAIGQASKQDVEAAYYRAVAKGAIIEGEARYTTTTDQRSKDAPPLPAMTKAEWQAQLTDKKGMNDSQA
KALVAEGIKSGRLKKTSHRVTTVEGIRVEKAVLNIEARGRGKVIAALTSEQVTVQLVGKTLKAEQQKSVV
DICTTTDRFVAAHGFAGTGKSYMTMSAKAVLESQGYNVTALAPYGTQKKSLEDEGMPARTVAAFVKAKDK
KIDEKSVVFIDEAGVIPARQMKVLMETIEKAGARAVFLGDTSQTKAVEAGKPFEQLISAGMQTSYMKDIQ
RQKNDVLLEAVKLAAEGHISGSLARLSNISAEKDQDKRLEAVANRWLSFSPEQREGTLIISGTNESRVIL
NSQIREALQLQGKGVEVNFLERVDSTQAERRDSKYYQVGQIIIPEKDYKNGLQRGESYRVLDTGPGNLLT
VAGSDGQKISFSPRTHKNLSVYTSVAAELAVGDKVMVTRNDKTLDVANGDRFTVTGTTEKGTFTLTNSKG
REIELDQKQAAYLSYAYATTVHKAQGLTCDRVLFNIDTRSLTTSKDVFYVGISRARHEVEIFTDDKKRLL
SSASRNSPKTTASEIDRFLGMEERYKDVNRSYGHSDGQQEKSYGHDAGHQTEEKENTMKNATEAAHDGPD
TSGTENTSQDLRQREQEEVNAAIRYEQRQEATPEMEANPYHDYERYADEGPDYGDMQDYSTYEEYERARD
VELPQHEHENHQHDHDKEGYGL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1185 | GenBank | WP_040223430 |
| Name | t4cp2_DL189_RS22765_PECIMP|punnamed2 |
UniProt ID | _ |
| Length | 510 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 510 a.a. Molecular weight: 57917.09 Da Isoelectric Point: 9.6871
>WP_040223430.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MTEQERGLIFLCSLLGPIPFVWFITAKFTYGINSETAHYLVPYLLKNTFNLWPLWAAIIAGVVIGLCGFI
GFIIYDKGRVYKGPRFKRILRGTKLARASTLAEKTKERGKKQLMIAGVPVPTEYETLHFSIGGATGTGKS
TVIKEAIYSSLKRGTDKRIVLDPNGEYVETFYRPGDVILSPYDKRSPGWSMFNEIRDDFDYDLYAVSIIQ
ESKERSAEEWYRYGRLIYREVSKKLLTLKRNPTMEEVMHWCTIVENDKLKEFVKGTPAEALFIKGAERAT
GSARFVLGDKLAPHLKMPAGDFSLRDWLEDDKPGTLFITWQEQMKKSLNPLISCWLDIMFSSLLGMGKRD
QRTLFFIDELESLEYLPNLQDLLTKGRKSGASVWAGYQTYSQLIERYGINIAETLLGSMRSGIVMGSSRL
GKETLEQMSRALGEIEGEVEHKQGNPFQAGIGNRPRIETRTVRAVTPTEISELKNLTGYVYFPGDLPVGK
FKTKYVKYTRKNPVPGIIMR
MTEQERGLIFLCSLLGPIPFVWFITAKFTYGINSETAHYLVPYLLKNTFNLWPLWAAIIAGVVIGLCGFI
GFIIYDKGRVYKGPRFKRILRGTKLARASTLAEKTKERGKKQLMIAGVPVPTEYETLHFSIGGATGTGKS
TVIKEAIYSSLKRGTDKRIVLDPNGEYVETFYRPGDVILSPYDKRSPGWSMFNEIRDDFDYDLYAVSIIQ
ESKERSAEEWYRYGRLIYREVSKKLLTLKRNPTMEEVMHWCTIVENDKLKEFVKGTPAEALFIKGAERAT
GSARFVLGDKLAPHLKMPAGDFSLRDWLEDDKPGTLFITWQEQMKKSLNPLISCWLDIMFSSLLGMGKRD
QRTLFFIDELESLEYLPNLQDLLTKGRKSGASVWAGYQTYSQLIERYGINIAETLLGSMRSGIVMGSSRL
GKETLEQMSRALGEIEGEVEHKQGNPFQAGIGNRPRIETRTVRAVTPTEISELKNLTGYVYFPGDLPVGK
FKTKYVKYTRKNPVPGIIMR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 15493..33002
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| DL189_RS22620 (DL189_22615) | 10936..11700 | + | 765 | WP_001389365 | IS6-like element IS6100 family transposase | - |
| DL189_RS25735 | 11840..11905 | - | 66 | Protein_13 | EAL domain-containing protein | - |
| DL189_RS22635 (DL189_22630) | 12110..12442 | + | 333 | WP_045723726 | hypothetical protein | - |
| DL189_RS22640 (DL189_22635) | 12636..12995 | + | 360 | WP_058672990 | hypothetical protein | - |
| DL189_RS22645 (DL189_22640) | 13000..13608 | + | 609 | WP_058672989 | recombinase family protein | - |
| DL189_RS22650 (DL189_22645) | 13605..13790 | - | 186 | WP_024562121 | ribbon-helix-helix domain-containing protein | - |
| DL189_RS22655 (DL189_22650) | 14075..14383 | - | 309 | WP_000733764 | hypothetical protein | - |
| DL189_RS22660 (DL189_22655) | 14387..14725 | - | 339 | WP_000040982 | hypothetical protein | - |
| DL189_RS22665 (DL189_22660) | 14722..15072 | - | 351 | WP_058672988 | hypothetical protein | - |
| DL189_RS22670 (DL189_22665) | 15076..15393 | - | 318 | WP_058672987 | H-NS family nucleoid-associated regulatory protein | - |
| DL189_RS22675 (DL189_22670) | 15493..16275 | + | 783 | WP_058672986 | lytic transglycosylase domain-containing protein | virB1 |
| DL189_RS22685 (DL189_22680) | 16522..16818 | + | 297 | WP_015060075 | TrbC/VirB2 family protein | virB2 |
| DL189_RS22690 (DL189_22685) | 16858..17175 | + | 318 | WP_040223409 | VirB3 family type IV secretion system protein | virB3 |
| DL189_RS22695 (DL189_22690) | 17176..19749 | + | 2574 | WP_040223410 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| DL189_RS22700 (DL189_22695) | 19765..20475 | + | 711 | WP_040223411 | type IV secretion system protein | virB5 |
| DL189_RS22705 (DL189_22700) | 20485..20727 | + | 243 | WP_040223412 | EexN family lipoprotein | - |
| DL189_RS22710 (DL189_22705) | 20744..21781 | + | 1038 | WP_058672985 | type IV secretion system protein | virB6 |
| DL189_RS26240 (DL189_22710) | 21858..21989 | + | 132 | WP_032493177 | hypothetical protein | - |
| DL189_RS22720 (DL189_22715) | 22000..22695 | + | 696 | WP_040223414 | type IV secretion system protein | virB8 |
| DL189_RS22725 (DL189_22720) | 22706..23593 | + | 888 | WP_040223415 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| DL189_RS22730 (DL189_22725) | 23593..24726 | + | 1134 | WP_040223416 | type IV secretion system protein VirB10 | virB10 |
| DL189_RS22735 (DL189_22730) | 24777..25778 | + | 1002 | WP_058672984 | ATPase, T2SS/T4P/T4SS family | virB11 |
| DL189_RS22740 (DL189_22735) | 25835..26302 | + | 468 | WP_050136493 | phospholipase D family protein | - |
| DL189_RS22745 (DL189_22740) | 26299..26610 | + | 312 | WP_040223418 | hypothetical protein | - |
| DL189_RS22750 (DL189_22745) | 27230..27592 | - | 363 | WP_024561906 | hypothetical protein | - |
| DL189_RS22755 (DL189_22750) | 27585..28250 | - | 666 | WP_040223419 | DUF6710 family protein | - |
| DL189_RS22760 (DL189_22755) | 28247..31465 | - | 3219 | WP_058672983 | MobF family relaxase | - |
| DL189_RS22765 (DL189_22760) | 31470..33002 | - | 1533 | WP_040223430 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| DL189_RS26060 | 33002..33427 | - | 426 | WP_227660646 | traK protein | - |
| DL189_RS22775 (DL189_22770) | 33895..34326 | + | 432 | WP_040223429 | plasmid stabilization protein StbA | - |
| DL189_RS22780 (DL189_22775) | 34335..35051 | + | 717 | WP_040223428 | StbB family protein | - |
| DL189_RS22785 (DL189_22780) | 35054..35422 | + | 369 | WP_040223427 | plasmid stabilization protein StbC | - |
| DL189_RS26065 | 35486..35836 | + | 351 | WP_227660645 | hypothetical protein | - |
| DL189_RS22795 (DL189_22790) | 35895..36077 | - | 183 | WP_040223426 | hypothetical protein | - |
| DL189_RS22800 (DL189_22795) | 36152..36358 | - | 207 | WP_048987680 | DUF905 family protein | - |
| DL189_RS25740 | 36539..37216 | - | 678 | WP_050136486 | DUF5417 domain-containing protein | - |
Host bacterium
| ID | 2241 | GenBank | NZ_QHMI01000031 |
| Plasmid name | PECIMP|punnamed2 | Incompatibility group | IncN3 |
| Plasmid size | 40047 bp | Coordinate of oriT [Strand] | 33573..33663 [+] |
| Host baterium | Enterobacter hormaechei strain PECIMP |
Cargo genes
| Drug resistance gene | blaIMP-1, dfrB3, cmlA1, qacE, sul1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |