Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101796 |
| Name | oriT_pSGL03 |
| Organism | Ligilactobacillus salivarius strain SGL 03 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_PECX01000002 (104542..104745 [+], 204 nt) |
| oriT length | 204 nt |
| IRs (inverted repeats) | 125..130, 143..148 (AAATCC..GGATTT) 66..72, 83..89 (ACCCCCC..GGGGGGT) |
| Location of nic site | 135..136 |
| Conserved sequence flanking the nic site |
TTTGGTTACA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 204 nt
>oriT_pSGL03
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
AAGCGGAAGTCGCAGGTGTGGACTGATCTTGCTGGCTGGTGTGGCAATAGCCACGCCAGCACTTAACCCCCCGTATCTAACAGGGGGGTACAAATCGACAGGAAACAGTCAAAAAAACATTAGAAAATCCTTTGGTTACAAGGGATTTACAAAATTTCAGCGTATGTCAAATGGGCTTTAAAAGTTGACATACGCCTTTTTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file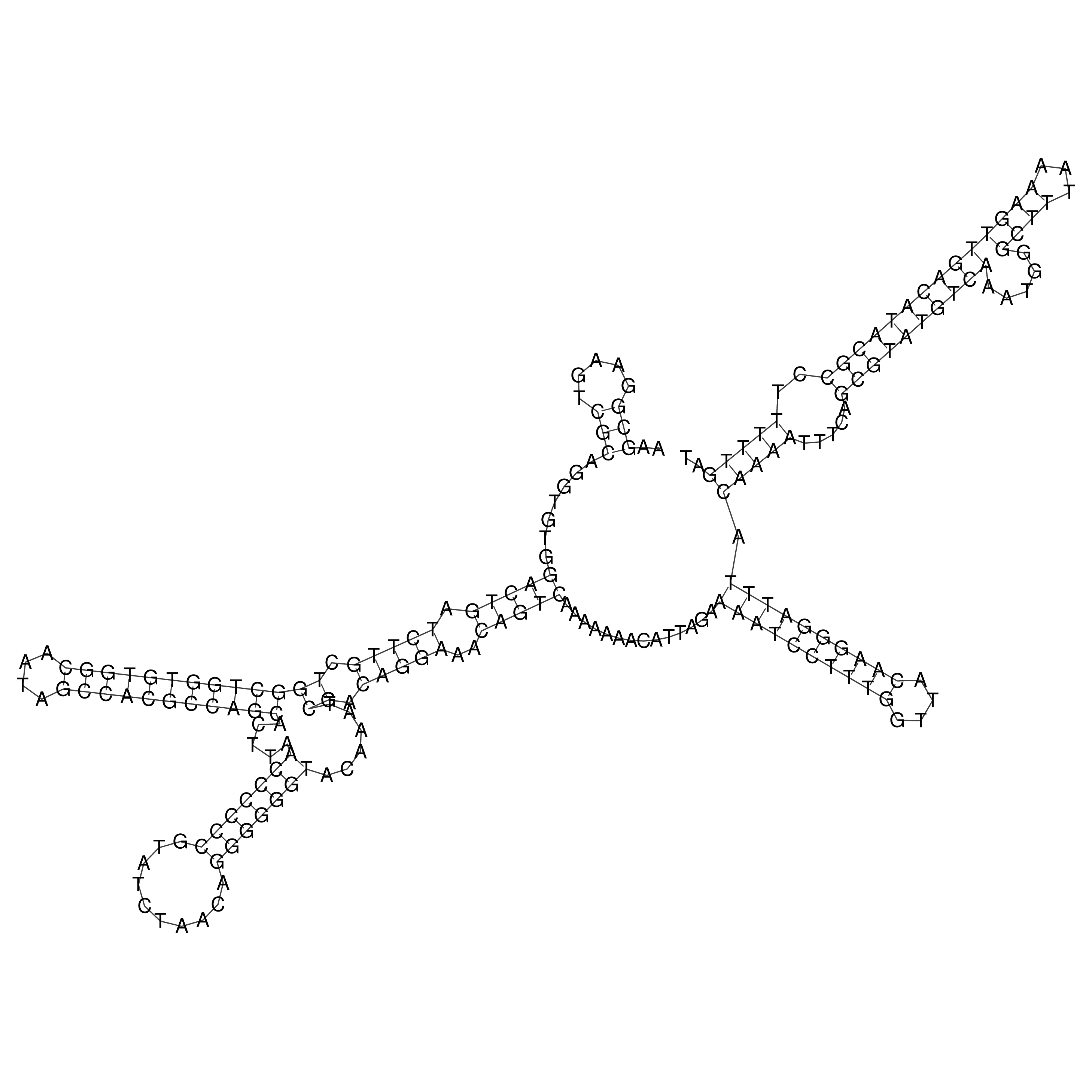
Relaxase
| ID | 1504 | GenBank | WP_000398284 |
| Name | mobT_CR166_RS09285_pSGL03 |
UniProt ID | A0A3P3PY43 |
| Length | 401 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 401 a.a. Molecular weight: 47398.10 Da Isoelectric Point: 6.5091
>WP_000398284.1 MULTISPECIES: MobT family relaxase [Bacteria]
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
MEGFLLNEQTWLQHLKEKRLAYGLSQNRLAVATGITRQYLSDIETGKVKPSEDLQQSLWEALERFNPDAP
LEMLFDYVRIRFPTTDVQQVVENILQLKLSYFLHEDYGFYSYSEHYALGDIFVLCSHELDKGVLVELKGR
GCRQFESYLLAQQRSWYEFFMDVLVAGGVMKRLDLAINDKTGILNIPVLTEKCQQEECISVFRSFKSYRS
GELVRKEEKECMGNTLYIGSLQSEVYFCIYEKDYEQYKKNDIPIEDAEVKNRFEIRLKNERAYYAVRDLL
VYDNPEHTAFKIINRYIRFVDKDDSKPRSDWKLNEEWAWFIGNNRERLKLTTKPEPYSFQRTLNWLSHQV
APTLKVAIKLDEINQTQVVKDILDHAKLTDRHKQILKQQSVKEQDVITTKK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A3P3PY43 |
Auxiliary protein
| ID | 604 | GenBank | WP_099460259 |
| Name | WP_099460259_pSGL03 |
UniProt ID | _ |
| Length | 405 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 405 a.a. Molecular weight: 47035.95 Da Isoelectric Point: 9.7978
>WP_099460259.1 tyrosine-type recombinase/integrase [Ligilactobacillus salivarius]
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDIVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
MSEKRRDNKGRILKTGESQRKDGRYLYKYIDSFGEPQFVYSWKLVATDRVPAGKRDCISLREKIAELQKD
IHDGIDIVGKKMTLCQLYAKQNAQRPKVRKNTETGRKYLMDILKKDKLGVRSIDSIKPSDAKEWAIRMSE
NGYAYQTINNYKRSLKASFYIAIQDDCVRKNPFDFQLKAVLDDDTVPKTVLTEEQEEKLLAFAKADKTYS
KNYDEILILLKTGLRISEFGGLTLPDLDFENRLVNIDHQLLRDTEIGYYIETPKTKSGERQVPMVEEAYQ
AFKRVLANRKNDKRVEIDGYSDFLFLNRKNYPKVASDYNGMMKGLVKKYNKYNEDKLPHITPHSLRHTFC
TNYANAGMNPKALQYIMGHANIAMTLNYYAHATFDSAMAEMKRLNKEKQQERLVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1184 | GenBank | WP_000813488 |
| Name | tcpA_CR166_RS09275_pSGL03 |
UniProt ID | _ |
| Length | 461 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 461 a.a. Molecular weight: 53370.27 Da Isoelectric Point: 9.0687
>WP_000813488.1 MULTISPECIES: FtsK/SpoIIIE domain-containing protein [Bacteria]
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
MKQRGKRIRPSGKDLVFHFTIASLLPVFLLVVGLFHVKTIQQINWQDFNLSQADKIDIPYLIISFSVAIL
ICLLVAFVFKRVRYDTVKQLYHRQKLAKMILENKWYESEQVKTEGFFKDSAGRTKEKITYFPKMYYRLKN
GLIQIRVEITLGKYQDQLLHLEKKLESGLYCELTDKELKDSYVEYTLLYDTIASRISIDEVEAKDGKLRL
MKNVWWEYDKLPHMLIAGGTGGGKTYFILTLIEALLHTDSKLYILDPKNADLADLGSVMANVYYRKEDLL
SCIETFYEEMMKRSEEMKQMKNYKTGKNYAYLGLPAHFLIFDEYVAFMEMLGTKENTAVMNKLKQIVMLG
RQAGFFLILACQRPDAKYLGDGIRDQFNFRVALGRMSEMGYGMMFGSDVQKDFFLKRIKGRGYVDVGTSV
ISEFYTPLVPKGYDFLEEIKKLSNSRQSTQATCEAEVAGVD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 102437..113847
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CR166_RS09235 (CR166_09220) | 98644..99045 | + | 402 | WP_242439822 | hypothetical protein | - |
| CR166_RS09240 (CR166_09225) | 99228..99569 | - | 342 | WP_003703433 | hypothetical protein | - |
| CR166_RS09245 (CR166_09230) | 99590..99856 | - | 267 | WP_160996539 | hypothetical protein | - |
| CR166_RS09250 (CR166_09235) | 100018..101196 | - | 1179 | WP_099460253 | pyridoxal phosphate-dependent aminotransferase | - |
| CR166_RS09255 (CR166_09240) | 101214..101999 | - | 786 | WP_099460254 | carbon-nitrogen family hydrolase | - |
| CR166_RS09265 (CR166_09250) | 102437..102751 | + | 315 | WP_000420682 | YdcP family protein | orf23 |
| CR166_RS09270 (CR166_09255) | 102767..103153 | + | 387 | WP_000985015 | YdcP family protein | orf23 |
| CR166_RS09275 (CR166_09260) | 103182..104567 | + | 1386 | WP_000813488 | FtsK/SpoIIIE domain-containing protein | virb4 |
| CR166_RS09280 (CR166_09265) | 104570..104722 | + | 153 | WP_000879507 | hypothetical protein | - |
| CR166_RS09285 (CR166_09270) | 104745..105950 | + | 1206 | WP_000398284 | MobT family relaxase | - |
| CR166_RS09290 (CR166_09275) | 105993..106214 | + | 222 | WP_001009056 | hypothetical protein | orf19 |
| CR166_RS09295 (CR166_09280) | 106331..106828 | + | 498 | WP_000342539 | antirestriction protein ArdA | - |
| CR166_RS09300 (CR166_09285) | 106917..107309 | + | 393 | WP_242439823 | conjugal transfer protein | orf17a |
| CR166_RS09305 (CR166_09290) | 107293..109740 | + | 2448 | WP_000331160 | ATP-binding protein | virb4 |
| CR166_RS09310 (CR166_09295) | 109743..111920 | + | 2178 | WP_099460256 | hypothetical protein | orf15 |
| CR166_RS09315 (CR166_09300) | 111917..112918 | + | 1002 | WP_099460257 | bifunctional lysozyme/C40 family peptidase | orf14 |
| CR166_RS09320 (CR166_09305) | 112915..113847 | + | 933 | WP_001224318 | conjugal transfer protein | orf13 |
| CR166_RS09325 (CR166_09310) | 114166..116085 | + | 1920 | WP_099460258 | tetracycline resistance ribosomal protection protein Tet(M) | - |
| CR166_RS09330 (CR166_09315) | 116204..116371 | + | 168 | WP_000336323 | cysteine-rich KTR domain-containing protein | - |
| CR166_RS09335 (CR166_09320) | 116431..116784 | - | 354 | WP_001227347 | helix-turn-helix transcriptional regulator | - |
| CR166_RS09345 (CR166_09325) | 117289..117711 | + | 423 | WP_000804885 | sigma-70 family RNA polymerase sigma factor | - |
| CR166_RS09350 (CR166_09330) | 117708..117938 | + | 231 | WP_000857133 | helix-turn-helix domain-containing protein | - |
| CR166_RS10525 (CR166_09335) | 118164..118415 | - | 252 | WP_001845478 | hypothetical protein | - |
| CR166_RS09360 (CR166_09340) | 118399..118602 | + | 204 | WP_000814511 | excisionase | - |
Host bacterium
| ID | 2240 | GenBank | NZ_PECX01000002 |
| Plasmid name | pSGL03 | Incompatibility group | - |
| Plasmid size | 175036 bp | Coordinate of oriT [Strand] | 104542..104745 [+] |
| Host baterium | Ligilactobacillus salivarius strain SGL 03 |
Cargo genes
| Drug resistance gene | tet(M) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIC1 |