Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101788 |
| Name | oriT_pRspCCGE510C |
| Organism | Rhizobium sp. CCGE 510 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AEYF01000048 (81169..81197 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRspCCGE510C
AGGGCGCAATATACGTCGCTGCCGCGACG
AGGGCGCAATATACGTCGCTGCCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file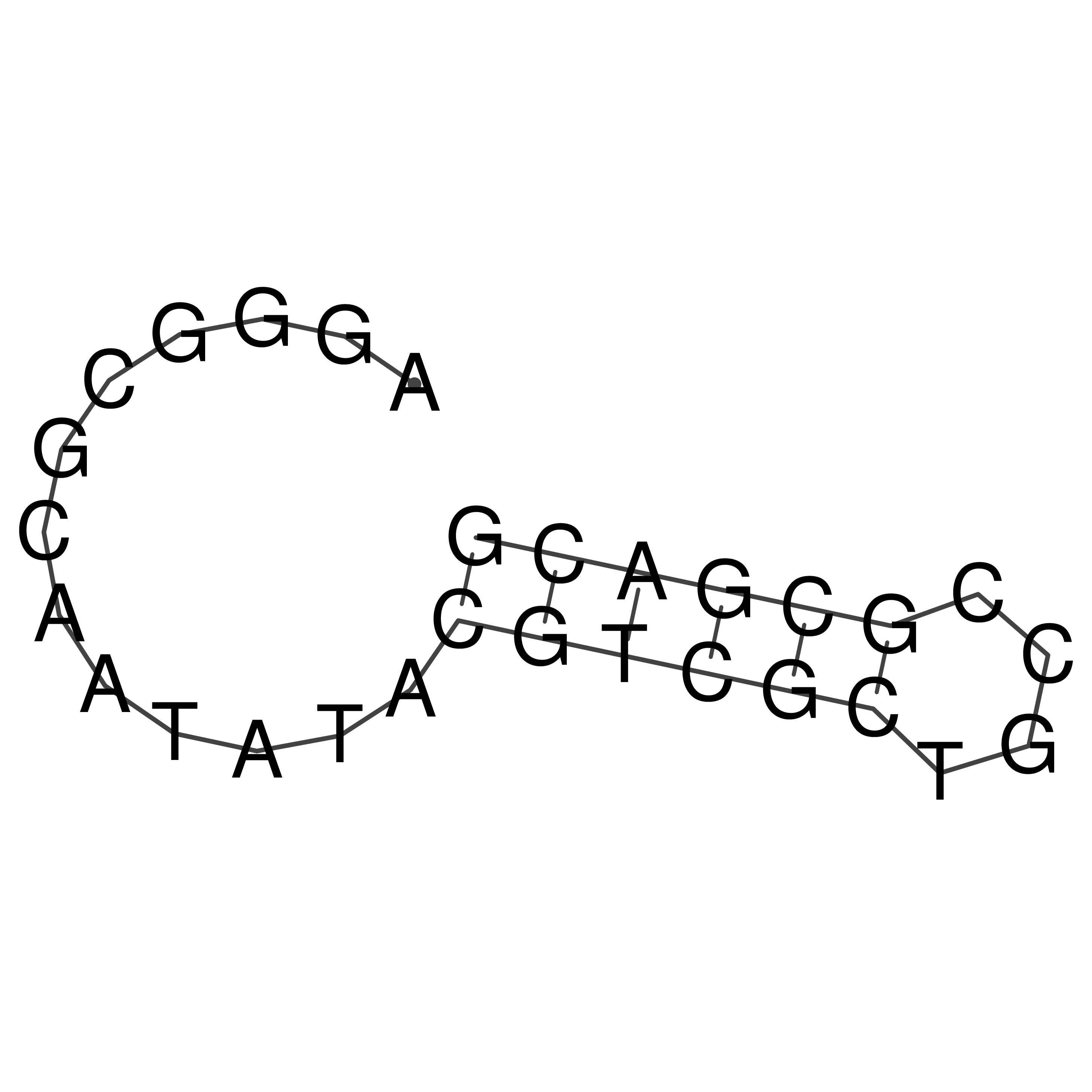
Relaxase
| ID | 1501 | GenBank | WP_007636131 |
| Name | traA_RCCGE510_RS28735_pRspCCGE510C |
UniProt ID | J5PM42 |
| Length | 1557 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1557 a.a. Molecular weight: 172019.37 Da Isoelectric Point: 7.1945
>WP_007636131.1 Ti-type conjugative transfer relaxase TraA [Rhizobium sp. CCGE 510]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDELAGTSFSYRGGAGELMHEEVALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENIALVREFVRDTLTSKGLIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVAVLGEGGKPLRVVTPDRPTGKIVYKVWAGDKETMKAWKIAWAEAANR
HLALAGHDIHLDGRSYAEQGLDGIAQKHLGPEKAALARKGKELYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALLRTVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSGRNVEAAMERVESSDPKTPFRLDAEQVDAVRHVTGDSGIAAVVGLAGAGKS
TLLAAARVAWESQGRRVMGAALAGKAAEGLQDSSGIRSRTLASWELAWANGRETLHRGDVVVIDEAGMVA
SQQMARVLKIAEEAEAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARDASRLFARGE
VEKGLDAYARHGHLVEAGTREETIDRIVSDWAVVRREAIERSTSEGRDGRLRGDELLVLAHTNDDVRKLN
QALRGVMTQEGALGESRSFRSERGVREFGAGDRIIFLENARFLEPRDRHAGPQYVKNGMLGTVVSTGDKR
DEVLLSVRLDNGSKVVFGEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRADDGTVHRLWGVSLPKA
LEEAGIQEGDTVMLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERESHRP
ELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAEKREQVA
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASVIGVAPRRLADDLAAAPGRLGRLRGSELI
VDGRAAREERNLAVAAVKELLPMARAHATEFRRNAERFELREQTRRAHMSLSIPTLSERAMARLVEIEAV
RSQGGDDAYKTAFALTAKDRSVVQEIKAVSEALTARFGWSAFSAKADVIAERNIVERMPEDLTDERRGKL
TPLFDAVRRFADEQHLAERRDRSKVVAGASADLSKEPGKEDVMPPMFAAVTEFKVPVDDEARSRALASPL
YRQRRAALAEAAGMVWRDPADFVDKVEDLLGKGFAADRIAAAVANDPAAYAALRGSDRLVDRMLASGRER
KEATRTVPEAAARLRALGAAYLNALDSERQAITDERRRMAVAIPALSKAAEEALAHLTVEVRKDSRKLSV
SAASLEQGIGREFAAVSRALDERFGRNALVRGDKDLVNRVPPPQRRAFAAMQERLKVLQQTVRLQSSEQI
IAERRQRAASRARGINL
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDELAGTSFSYRGGAGELMHEEVALPDEIPDWLRSAISG
QSVSKASEVFWNAVDAFETRADAQLARELIIALPEELTRAENIALVREFVRDTLTSKGLIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGAKKVAVLGEGGKPLRVVTPDRPTGKIVYKVWAGDKETMKAWKIAWAEAANR
HLALAGHDIHLDGRSYAEQGLDGIAQKHLGPEKAALARKGKELYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALLRTVDDPTDFANIRARLMASDQLVILKPQEIEAETGKVSEPAVFTTREMLRIE
YDMAQSARVLSERRGFGVSGRNVEAAMERVESSDPKTPFRLDAEQVDAVRHVTGDSGIAAVVGLAGAGKS
TLLAAARVAWESQGRRVMGAALAGKAAEGLQDSSGIRSRTLASWELAWANGRETLHRGDVVVIDEAGMVA
SQQMARVLKIAEEAEAKVVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQREAWARDASRLFARGE
VEKGLDAYARHGHLVEAGTREETIDRIVSDWAVVRREAIERSTSEGRDGRLRGDELLVLAHTNDDVRKLN
QALRGVMTQEGALGESRSFRSERGVREFGAGDRIIFLENARFLEPRDRHAGPQYVKNGMLGTVVSTGDKR
DEVLLSVRLDNGSKVVFGEDSYRNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRVDL
YAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADVRADDGTVHRLWGVSLPKA
LEEAGIQEGDTVMLRKDGVERVKVQIAVVDEKTGHKHYEEREVDRNVWTASQVETASARQERIERESHRP
ELFNPLVERLSRSGAKTTTLDFESEASYRAHANDFARRRGLDHLSLAAAEMEESLTRRWAWIAEKREQVA
KLWERASVALGFAIERERRVAYNEERSQTMVEATSSDARTASHSVSGASAAETRYLIPPATSFVSSVEED
ARLAQLASPAWTEREVILRPLLQKIYRDPDAALVSLNALASVIGVAPRRLADDLAAAPGRLGRLRGSELI
VDGRAAREERNLAVAAVKELLPMARAHATEFRRNAERFELREQTRRAHMSLSIPTLSERAMARLVEIEAV
RSQGGDDAYKTAFALTAKDRSVVQEIKAVSEALTARFGWSAFSAKADVIAERNIVERMPEDLTDERRGKL
TPLFDAVRRFADEQHLAERRDRSKVVAGASADLSKEPGKEDVMPPMFAAVTEFKVPVDDEARSRALASPL
YRQRRAALAEAAGMVWRDPADFVDKVEDLLGKGFAADRIAAAVANDPAAYAALRGSDRLVDRMLASGRER
KEATRTVPEAAARLRALGAAYLNALDSERQAITDERRRMAVAIPALSKAAEEALAHLTVEVRKDSRKLSV
SAASLEQGIGREFAAVSRALDERFGRNALVRGDKDLVNRVPPPQRRAFAAMQERLKVLQQTVRLQSSEQI
IAERRQRAASRARGINL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1180 | GenBank | WP_007636149 |
| Name | traG_RCCGE510_RS28750_pRspCCGE510C |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70743.73 Da Isoelectric Point: 9.4898
>WP_007636149.1 Ti-type conjugative transfer system protein TraG [Rhizobium sp. CCGE 510]
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKTEYWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLFCFLGVAAFYALREFGRLAPPVQARVVSWDRALSYLDMVAVIAAVAGFMAVAVSARI
SVVVPDEIKRARRGIFGDADWLSMAAARKLFPPDGEIVVGERYRVDKEIVHELPFDANDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPMICLDPSTEVAPMVLEHRTNALGREVMVLDPT
NPTTGFNVLDGIETSSQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GKTFKSRDIVSGRKDVFLNIPAPILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMMMYQSVGQLERHFGKDGATSWIEGCAFASYAAIKALDTARSVSAQCGEMTVEVKG
SSRNLGWDNRTGGTRKSESISFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKDMDAAAK
ANRFVKSIT
MGLRGKPHPSLLLVLVPIAVTTITIYIVGWRWPGLAAGMSGKTEYWFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLFCFLGVAAFYALREFGRLAPPVQARVVSWDRALSYLDMVAVIAAVAGFMAVAVSARI
SVVVPDEIKRARRGIFGDADWLSMAAARKLFPPDGEIVVGERYRVDKEIVHELPFDANDRSTWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPMICLDPSTEVAPMVLEHRTNALGREVMVLDPT
NPTTGFNVLDGIETSSQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLAMLRDIQENSQSDFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GKTFKSRDIVSGRKDVFLNIPAPILRSYPGIGRVIIGSLINAMMQADGAFRRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMMMYQSVGQLERHFGKDGATSWIEGCAFASYAAIKALDTARSVSAQCGEMTVEVKG
SSRNLGWDNRTGGTRKSESISFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKDMDAAAK
ANRFVKSIT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 81824..99818
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RCCGE510_RS28740 (RCCGE510_29091) | 81317..81604 | + | 288 | WP_007636139 | TraC family protein | - |
| RCCGE510_RS28745 (RCCGE510_29096) | 81594..81833 | + | 240 | WP_007636148 | conjugal transfer protein TraD | - |
| RCCGE510_RS28750 (RCCGE510_29101) | 81824..83743 | + | 1920 | WP_007636149 | Ti-type conjugative transfer system protein TraG | virb4 |
| RCCGE510_RS28755 (RCCGE510_29106) | 83779..84193 | - | 415 | Protein_91 | hypothetical protein | - |
| RCCGE510_RS28760 (RCCGE510_29111) | 84193..84459 | - | 267 | WP_007636151 | type II toxin-antitoxin system prevent-host-death family antitoxin | - |
| RCCGE510_RS28765 (RCCGE510_29116) | 84608..85078 | - | 471 | WP_007636152 | hypothetical protein | - |
| RCCGE510_RS35310 (RCCGE510_29121) | 85402..85569 | - | 168 | WP_164841435 | hypothetical protein | - |
| RCCGE510_RS28775 (RCCGE510_29126) | 85889..86206 | - | 318 | WP_007636154 | DUF736 domain-containing protein | - |
| RCCGE510_RS33930 (RCCGE510_29136) | 86847..87104 | - | 258 | WP_037080160 | hypothetical protein | - |
| RCCGE510_RS28790 (RCCGE510_29141) | 87463..88653 | + | 1191 | WP_007636159 | DUF1173 domain-containing protein | - |
| RCCGE510_RS28795 (RCCGE510_29146) | 89089..89610 | + | 522 | WP_037080293 | excisionase | - |
| RCCGE510_RS28800 (RCCGE510_29151) | 89600..90178 | + | 579 | WP_007636163 | PIN domain-containing protein | - |
| RCCGE510_RS28805 (RCCGE510_29156) | 90222..91250 | - | 1029 | WP_007636165 | P-type DNA transfer ATPase VirB11 | virB11 |
| RCCGE510_RS28810 (RCCGE510_29161) | 91259..92431 | - | 1173 | WP_007636166 | type IV secretion system protein VirB10 | virB10 |
| RCCGE510_RS28815 (RCCGE510_29166) | 92440..93297 | - | 858 | WP_007636174 | P-type conjugative transfer protein VirB9 | virB9 |
| RCCGE510_RS28820 (RCCGE510_29171) | 93294..93965 | - | 672 | WP_007636176 | virB8 family protein | virB8 |
| RCCGE510_RS35740 | 93967..94248 | - | 282 | WP_081525799 | hypothetical protein | - |
| RCCGE510_RS28825 (RCCGE510_29176) | 94288..95220 | - | 933 | WP_007636177 | type IV secretion system protein | virB6 |
| RCCGE510_RS28830 (RCCGE510_29181) | 95223..95456 | - | 234 | WP_007636178 | EexN family lipoprotein | - |
| RCCGE510_RS28835 (RCCGE510_29186) | 95453..96154 | - | 702 | WP_007636183 | P-type DNA transfer protein VirB5 | virB5 |
| RCCGE510_RS28840 (RCCGE510_29191) | 96151..98517 | - | 2367 | WP_007636185 | VirB4 family type IV secretion system protein | virb4 |
| RCCGE510_RS28845 (RCCGE510_29196) | 98510..98848 | - | 339 | WP_007636187 | type IV secretion system protein VirB3 | virB3 |
| RCCGE510_RS28850 (RCCGE510_29201) | 98854..99153 | - | 300 | WP_007636189 | TrbC/VirB2 family protein | virB2 |
| RCCGE510_RS28855 (RCCGE510_29206) | 99150..99818 | - | 669 | WP_007636191 | transglycosylase SLT domain-containing protein | virB1 |
| RCCGE510_RS28860 (RCCGE510_29211) | 99822..100358 | - | 537 | WP_007636192 | hypothetical protein | - |
| RCCGE510_RS28865 (RCCGE510_29216) | 100457..100828 | + | 372 | WP_007636193 | hypothetical protein | - |
| RCCGE510_RS34910 | 101683..101922 | - | 240 | WP_037080298 | hypothetical protein | - |
| RCCGE510_RS34915 | 102023..102306 | - | 284 | Protein_115 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| RCCGE510_RS28880 (RCCGE510_29226) | 102293..102592 | - | 300 | WP_007636195 | type II toxin-antitoxin system RelB/DinJ family antitoxin | - |
| RCCGE510_RS28885 | 103413..104605 | - | 1193 | Protein_117 | IS256 family transposase | - |
Host bacterium
| ID | 2232 | GenBank | NZ_AEYF01000048 |
| Plasmid name | pRspCCGE510C | Incompatibility group | - |
| Plasmid size | 393475 bp | Coordinate of oriT [Strand] | 81169..81197 [-] |
| Host baterium | Rhizobium sp. CCGE 510 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | hsiC1/vipB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |