Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101761 |
| Name | oriT_pRHBSTW-00215_3 |
| Organism | Klebsiella sp. RHBSTW-00215 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JABXQS010000003 (84976..85025 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pRHBSTW-00215_3
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file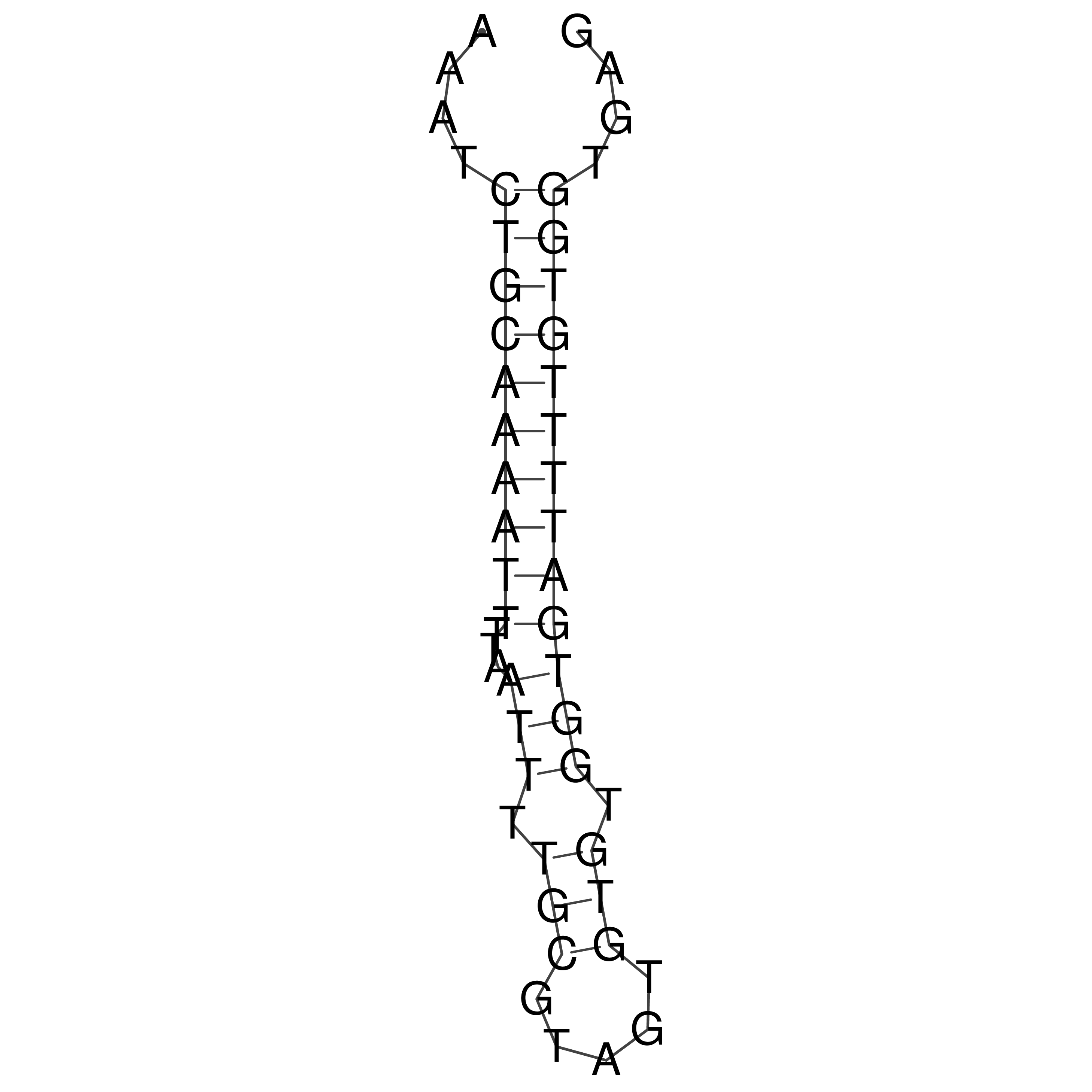
Relaxase
| ID | 1484 | GenBank | WP_082226159 |
| Name | traI_HV127_RS31140_pRHBSTW-00215_3 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190416.67 Da Isoelectric Point: 5.5084
>WP_082226159.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTNNLI
IARFNHDTSRAQEPQIHTHSVVINATQNGDKWQTLGSDTVGKTGFSENILANRIAFGKIYQSTLRADVES
MGYKTVDAGKHGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADTRAKELASAPAAPVKTDVPDISDVVTKAIAGLSDRKVQFTYADLLARTVSQLEA
KPGVFEQTRTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDTAVVRQSPF
SDAVSVLAQDRPTMGIVSGQGGAAGQRERVAELTLMAREQGRDVHIIAADNRSREFLSADARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKMSLKETLSMLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGKQATADIISEPEKGARYSRLAQEFAASVREGQESVAQISGSREQGILNGLIRETLKSEGVLGEKDM
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRQHDRFVIDRVTASSNMLTLRDKEGERLDMKVSAVD
SQWTLFRADKLTVAEGERLAVLGKIPDTRLKGGESVTVLKAEEGQLTVQRPGQKTTQSLSVGSSPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQGTARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIASQKAGLHTPAEQVIHLSIPLLESEELTFTRPQLLATALETGGGKVPMTEIDATIQ
SQIQAGSLLNVPVAPGHGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDNVPGSLMTDLTSGQRASTRM
ILETTDRFTVVQGYAGVGKTTQFRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVNRALATIEQVTPEQVPRKDGAWVPENSVVEFTPAQEEAIQK
ALNKGESVPAGQPTTLYEALVKDYADRTPEAQSQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKSTRTLNPKADQAQQHIDLAYAITA
HGAQGASEPFAIALEGVAGGRERMASFESAYVGLSRMKQHVQVYTDNREGWIKAINNSPEKATAHDILEP
RNDRAVKTAGQLFSRARALDEAAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDNDGRLQVIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMERQTEQATREVSGEEKKAGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVVSESLQRDRLQQMERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTNNLI
IARFNHDTSRAQEPQIHTHSVVINATQNGDKWQTLGSDTVGKTGFSENILANRIAFGKIYQSTLRADVES
MGYKTVDAGKHGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADTRAKELASAPAAPVKTDVPDISDVVTKAIAGLSDRKVQFTYADLLARTVSQLEA
KPGVFEQTRTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEIQRQNHVSVTPDTAVVRQSPF
SDAVSVLAQDRPTMGIVSGQGGAAGQRERVAELTLMAREQGRDVHIIAADNRSREFLSADARLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKMSLKETLSMLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGKQATADIISEPEKGARYSRLAQEFAASVREGQESVAQISGSREQGILNGLIRETLKSEGVLGEKDM
TVTALTPVWLDSKSRGVRDYYREGMVMERWDPETRQHDRFVIDRVTASSNMLTLRDKEGERLDMKVSAVD
SQWTLFRADKLTVAEGERLAVLGKIPDTRLKGGESVTVLKAEEGQLTVQRPGQKTTQSLSVGSSPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQGTARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIASQKAGLHTPAEQVIHLSIPLLESEELTFTRPQLLATALETGGGKVPMTEIDATIQ
SQIQAGSLLNVPVAPGHGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDNVPGSLMTDLTSGQRASTRM
ILETTDRFTVVQGYAGVGKTTQFRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVNRALATIEQVTPEQVPRKDGAWVPENSVVEFTPAQEEAIQK
ALNKGESVPAGQPTTLYEALVKDYADRTPEAQSQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKSTRTLNPKADQAQQHIDLAYAITA
HGAQGASEPFAIALEGVAGGRERMASFESAYVGLSRMKQHVQVYTDNREGWIKAINNSPEKATAHDILEP
RNDRAVKTAGQLFSRARALDEAAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDNDGRLQVIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMERQTEQATREVSGEEKKAGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVVSESLQRDRLQQMERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 594 | GenBank | WP_053390193 |
| Name | WP_053390193_pRHBSTW-00215_3 |
UniProt ID | A0A8G2A327 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 14883.89 Da Isoelectric Point: 4.3779
>WP_053390193.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacteriaceae]
MAKVQAYVSDEVADKINAIVEKRRVEGAKDKDISFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LESVIKTQFTVNKVLGIECLSPHVSGNPRWEWPGLIENIRDDVQEVMLRFFPDEESEDEE
MAKVQAYVSDEVADKINAIVEKRRVEGAKDKDISFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LESVIKTQFTVNKVLGIECLSPHVSGNPRWEWPGLIENIRDDVQEVMLRFFPDEESEDEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1159 | GenBank | WP_064386203 |
| Name | traD_HV127_RS31135_pRHBSTW-00215_3 |
UniProt ID | _ |
| Length | 772 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 772 a.a. Molecular weight: 86585.38 Da Isoelectric Point: 5.0283
>WP_064386203.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella/Raoultella group]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGKSLEYNSEQILADKYTIWCGEQLWTSFVFAAIVSLTVCIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKHSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDIWRECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFSEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLKNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLGMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDVQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRNLDTRVDARLNALLEAREAEGSLARTLFTPDAPAPELAEKDEKVGNPPEQSQPAE
SSGTSATVKVPSTAKTPEADESGQSPEVSNPAALLTKVVTVPLTRPRPAAATGTAAVASATDVPATPAGG
TEQALETQPAEQGQDMLPPGVNEYGEIDDMHAWDEWQSGEQTQRDMQRREEVNINHSHRRDEQDDIEIGG
NF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFIIFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGKSLEYNSEQILADKYTIWCGEQLWTSFVFAAIVSLTVCIVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKHSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDIWRECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFSEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLKNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLGMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDVQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRNLDTRVDARLNALLEAREAEGSLARTLFTPDAPAPELAEKDEKVGNPPEQSQPAE
SSGTSATVKVPSTAKTPEADESGQSPEVSNPAALLTKVVTVPLTRPRPAAATGTAAVASATDVPATPAGG
TEQALETQPAEQGQDMLPPGVNEYGEIDDMHAWDEWQSGEQTQRDMQRREEVNINHSHRRDEQDDIEIGG
NF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 87437..113426
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV127_RS30955 (HV127_30950) | 83844..84386 | + | 543 | WP_053390246 | antirestriction protein | - |
| HV127_RS30960 (HV127_30955) | 84409..84831 | - | 423 | WP_077268549 | transglycosylase SLT domain-containing protein | - |
| HV127_RS30965 (HV127_30960) | 85337..85729 | + | 393 | WP_053390193 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV127_RS30970 (HV127_30965) | 85967..86668 | + | 702 | WP_094898581 | hypothetical protein | - |
| HV127_RS30975 (HV127_30970) | 86829..86990 | + | 162 | WP_172686697 | TraY domain-containing protein | - |
| HV127_RS30980 (HV127_30975) | 87055..87423 | + | 369 | WP_064386081 | type IV conjugative transfer system pilin TraA | - |
| HV127_RS30985 (HV127_30980) | 87437..87742 | + | 306 | WP_032744112 | type IV conjugative transfer system protein TraL | traL |
| HV127_RS30990 (HV127_30985) | 87761..88327 | + | 567 | WP_064386078 | type IV conjugative transfer system protein TraE | traE |
| HV127_RS30995 (HV127_30990) | 88314..89048 | + | 735 | WP_182240935 | type-F conjugative transfer system secretin TraK | traK |
| HV127_RS31000 (HV127_30995) | 89048..90472 | + | 1425 | WP_064386076 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV127_RS31005 (HV127_31000) | 90535..91104 | + | 570 | WP_074182910 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV127_RS31010 (HV127_31005) | 91218..91496 | + | 279 | WP_064386123 | hypothetical protein | - |
| HV127_RS31015 (HV127_31010) | 91503..91745 | + | 243 | WP_064386074 | hypothetical protein | - |
| HV127_RS31020 (HV127_31015) | 91742..92146 | + | 405 | WP_064386072 | hypothetical protein | - |
| HV127_RS31025 (HV127_31020) | 92143..92490 | + | 348 | WP_064386070 | hypothetical protein | - |
| HV127_RS31030 (HV127_31025) | 92505..92885 | + | 381 | WP_100663260 | hypothetical protein | - |
| HV127_RS31035 (HV127_31030) | 92969..95608 | + | 2640 | WP_182240937 | type IV secretion system protein TraC | virb4 |
| HV127_RS31040 (HV127_31035) | 95608..96000 | + | 393 | WP_064343534 | type-F conjugative transfer system protein TrbI | - |
| HV127_RS31045 (HV127_31040) | 95997..96623 | + | 627 | WP_064343535 | type-F conjugative transfer system protein TraW | traW |
| HV127_RS31050 (HV127_31045) | 96667..97626 | + | 960 | WP_229295972 | conjugal transfer pilus assembly protein TraU | traU |
| HV127_RS31055 (HV127_31050) | 97639..98286 | + | 648 | WP_064343536 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV127_RS31060 (HV127_31055) | 98283..98666 | + | 384 | WP_053390297 | hypothetical protein | - |
| HV127_RS31065 (HV127_31060) | 98663..100618 | + | 1956 | WP_064359956 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV127_RS31070 (HV127_31065) | 100650..100910 | + | 261 | WP_053390216 | hypothetical protein | - |
| HV127_RS31075 (HV127_31070) | 100903..101514 | + | 612 | WP_053390217 | hypothetical protein | - |
| HV127_RS31080 (HV127_31075) | 101504..101746 | + | 243 | WP_053390218 | conjugal transfer protein TrbE | - |
| HV127_RS31085 (HV127_31080) | 101792..102118 | + | 327 | WP_053390219 | hypothetical protein | - |
| HV127_RS31090 (HV127_31085) | 102139..102891 | + | 753 | WP_053390220 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV127_RS31095 (HV127_31090) | 102902..103138 | + | 237 | WP_182240939 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV127_RS31100 (HV127_31095) | 103113..103679 | + | 567 | WP_100663597 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV127_RS31105 (HV127_31100) | 103672..104100 | + | 429 | WP_100663598 | conjugal transfer protein TrbF | - |
| HV127_RS31110 (HV127_31105) | 104087..105457 | + | 1371 | WP_182240941 | conjugal transfer pilus assembly protein TraH | traH |
| HV127_RS31115 (HV127_31110) | 105457..108336 | + | 2880 | WP_064386194 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV127_RS31120 (HV127_31115) | 108353..109021 | + | 669 | WP_128317877 | hypothetical protein | - |
| HV127_RS31125 (HV127_31120) | 109374..110105 | + | 732 | WP_064386198 | conjugal transfer complement resistance protein TraT | - |
| HV127_RS31945 | 110297..110989 | + | 693 | WP_202395549 | hypothetical protein | - |
| HV127_RS31135 (HV127_31130) | 111108..113426 | + | 2319 | WP_064386203 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2205 | GenBank | NZ_JABXQS010000003 |
| Plasmid name | pRHBSTW-00215_3 | Incompatibility group | IncFII |
| Plasmid size | 124557 bp | Coordinate of oriT [Strand] | 84976..85025 [-] |
| Host baterium | Klebsiella sp. RHBSTW-00215 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |