Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101757 |
| Name | oriT_pRHBSTW-00092_2 |
| Organism | Klebsiella grimontii strain RHBSTW-00092 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JABXRN010000002 (171796..171845 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pRHBSTW-00092_2
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file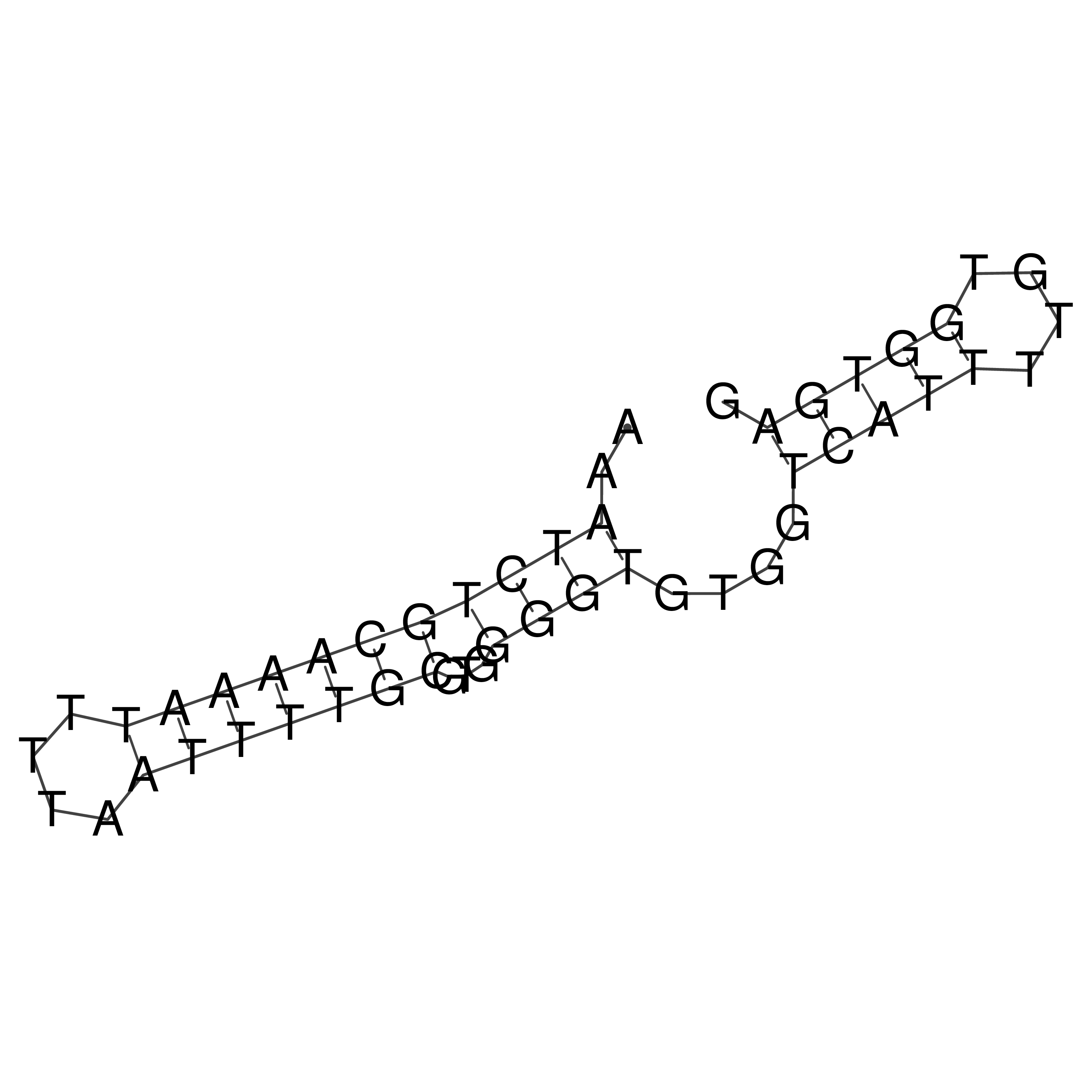
Relaxase
| ID | 1480 | GenBank | WP_065953842 |
| Name | traI_HV064_RS27895_pRHBSTW-00092_2 |
UniProt ID | A0A839CRE9 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190420.61 Da Isoelectric Point: 6.3863
>WP_065953842.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MMSIGSVKSADKAADYYTNKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADSRAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGLESVAQISGTREQSVLNGLIRDSLRNEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMESWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPAQEKAIQ
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHGARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQMITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARLLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
VDRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSADKAADYYTNKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADSRAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGLESVAQISGTREQSVLNGLIRDSLRNEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMESWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPAQEKAIQ
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHGARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQMITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARLLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
VDRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1155 | GenBank | WP_032436737 |
| Name | traD_HV064_RS27900_pRHBSTW-00092_2 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85553.49 Da Isoelectric Point: 5.0065
>WP_032436737.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREGRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGERPELASQPA
PAEVTVSPAPVKAPATTTMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREGRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGERPELASQPA
PAEVTVSPAPVKAPATTTMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 142590..172403
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV064_RS27900 (HV064_27895) | 142590..144899 | - | 2310 | WP_032436737 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HV064_RS30515 | 144955..145068 | - | 114 | Protein_145 | hypothetical protein | - |
| HV064_RS30710 | 145259..145438 | - | 180 | WP_317872608 | hypothetical protein | - |
| HV064_RS27905 (HV064_27900) | 145420..146400 | - | 981 | WP_014386491 | IS5-like element ISKpn26 family transposase | - |
| HV064_RS27910 (HV064_27905) | 146441..147190 | - | 750 | WP_182189496 | conjugal transfer complement resistance protein TraT | - |
| HV064_RS27915 (HV064_27910) | 147442..148044 | - | 603 | WP_023280878 | hypothetical protein | - |
| HV064_RS27920 (HV064_27915) | 148068..150890 | - | 2823 | WP_023320108 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV064_RS27925 (HV064_27920) | 150890..152269 | - | 1380 | WP_182189498 | conjugal transfer pilus assembly protein TraH | traH |
| HV064_RS27930 (HV064_27925) | 152247..152690 | - | 444 | WP_182189500 | F-type conjugal transfer protein TrbF | - |
| HV064_RS27935 (HV064_27930) | 152735..153292 | - | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV064_RS27940 (HV064_27935) | 153264..153503 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV064_RS27945 (HV064_27940) | 153514..154266 | - | 753 | WP_015065637 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV064_RS27950 (HV064_27945) | 154287..154613 | - | 327 | WP_032436744 | hypothetical protein | - |
| HV064_RS30715 | 154659..154844 | - | 186 | WP_032436745 | hypothetical protein | - |
| HV064_RS27955 (HV064_27950) | 154841..155077 | - | 237 | WP_032436746 | conjugal transfer protein TrbE | - |
| HV064_RS27960 (HV064_27955) | 155109..157064 | - | 1956 | WP_023307537 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV064_RS27965 (HV064_27960) | 157123..157761 | - | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV064_RS27970 (HV064_27965) | 157774..158733 | - | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| HV064_RS27975 (HV064_27970) | 158777..159403 | - | 627 | WP_023307538 | type-F conjugative transfer system protein TraW | traW |
| HV064_RS27980 (HV064_27975) | 159403..159792 | - | 390 | WP_032436749 | type-F conjugative transfer system protein TrbI | - |
| HV064_RS27985 (HV064_27980) | 159792..162431 | - | 2640 | WP_016528865 | type IV secretion system protein TraC | virb4 |
| HV064_RS27990 (HV064_27985) | 162503..162901 | - | 399 | WP_072158599 | hypothetical protein | - |
| HV064_RS27995 (HV064_27990) | 162909..163199 | - | 291 | WP_023307542 | hypothetical protein | - |
| HV064_RS28000 (HV064_27995) | 163196..163600 | - | 405 | WP_023307543 | hypothetical protein | - |
| HV064_RS28005 (HV064_28000) | 163645..163983 | - | 339 | WP_182189501 | hypothetical protein | - |
| HV064_RS28010 (HV064_28005) | 163984..164202 | - | 219 | WP_004171484 | hypothetical protein | - |
| HV064_RS28015 (HV064_28010) | 164226..164516 | - | 291 | WP_032423381 | hypothetical protein | - |
| HV064_RS28020 (HV064_28015) | 164521..164931 | - | 411 | WP_023307546 | hypothetical protein | - |
| HV064_RS28025 (HV064_28020) | 165063..165632 | - | 570 | WP_023307547 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV064_RS28030 (HV064_28025) | 165844..166263 | - | 420 | Protein_173 | conjugal transfer pilus-stabilizing protein TraP | - |
| HV064_RS28035 (HV064_28030) | 166256..167680 | - | 1425 | WP_023307549 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV064_RS28040 (HV064_28035) | 167680..168420 | - | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| HV064_RS28045 (HV064_28040) | 168407..168973 | - | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| HV064_RS28050 (HV064_28045) | 168993..169298 | - | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| HV064_RS28055 (HV064_28050) | 169312..169680 | - | 369 | WP_023307550 | type IV conjugative transfer system pilin TraA | - |
| HV064_RS28060 (HV064_28055) | 169734..170105 | - | 372 | WP_004208838 | TraY domain-containing protein | - |
| HV064_RS28065 (HV064_28060) | 170189..170884 | - | 696 | WP_023307551 | transcriptional regulator TraJ family protein | - |
| HV064_RS28070 (HV064_28065) | 171091..171483 | - | 393 | WP_032436755 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV064_RS28075 (HV064_28070) | 171918..172403 | + | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| HV064_RS28080 (HV064_28075) | 172436..172750 | - | 315 | WP_148673127 | DUF5983 family protein | - |
| HV064_RS28085 (HV064_28080) | 172798..173619 | - | 822 | WP_004182076 | DUF932 domain-containing protein | - |
| HV064_RS28090 (HV064_28085) | 174448..174861 | - | 414 | WP_023280875 | transcriptional regulator | - |
| HV064_RS28095 (HV064_28090) | 174862..175140 | - | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| HV064_RS28100 (HV064_28095) | 175130..175450 | - | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HV064_RS28105 (HV064_28100) | 175531..175755 | - | 225 | WP_004152719 | hypothetical protein | - |
| HV064_RS28110 (HV064_28105) | 175766..175978 | - | 213 | WP_004152718 | hypothetical protein | - |
| HV064_RS28115 (HV064_28110) | 176039..176395 | - | 357 | WP_004152717 | hypothetical protein | - |
| HV064_RS28120 (HV064_28115) | 177032..177382 | - | 351 | WP_004152758 | hypothetical protein | - |
Host bacterium
| ID | 2201 | GenBank | NZ_JABXRN010000002 |
| Plasmid name | pRHBSTW-00092_2 | Incompatibility group | IncFIB |
| Plasmid size | 208049 bp | Coordinate of oriT [Strand] | 171796..171845 [+] |
| Host baterium | Klebsiella grimontii strain RHBSTW-00092 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | mrkA, mrkB, mrkC, mrkD, mrkF, mrkJ |
| Metal resistance gene | arsC, arsB, arsA, arsD, arsR, silP, silA, silB, silF, silC, silR, silS, silE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |