Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101743 |
| Name | oriT_pRHBSTW-00189_4 |
| Organism | Klebsiella grimontii strain RHBSTW-00189 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JABXQY010000004 (74322..74370 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pRHBSTW-00189_4
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file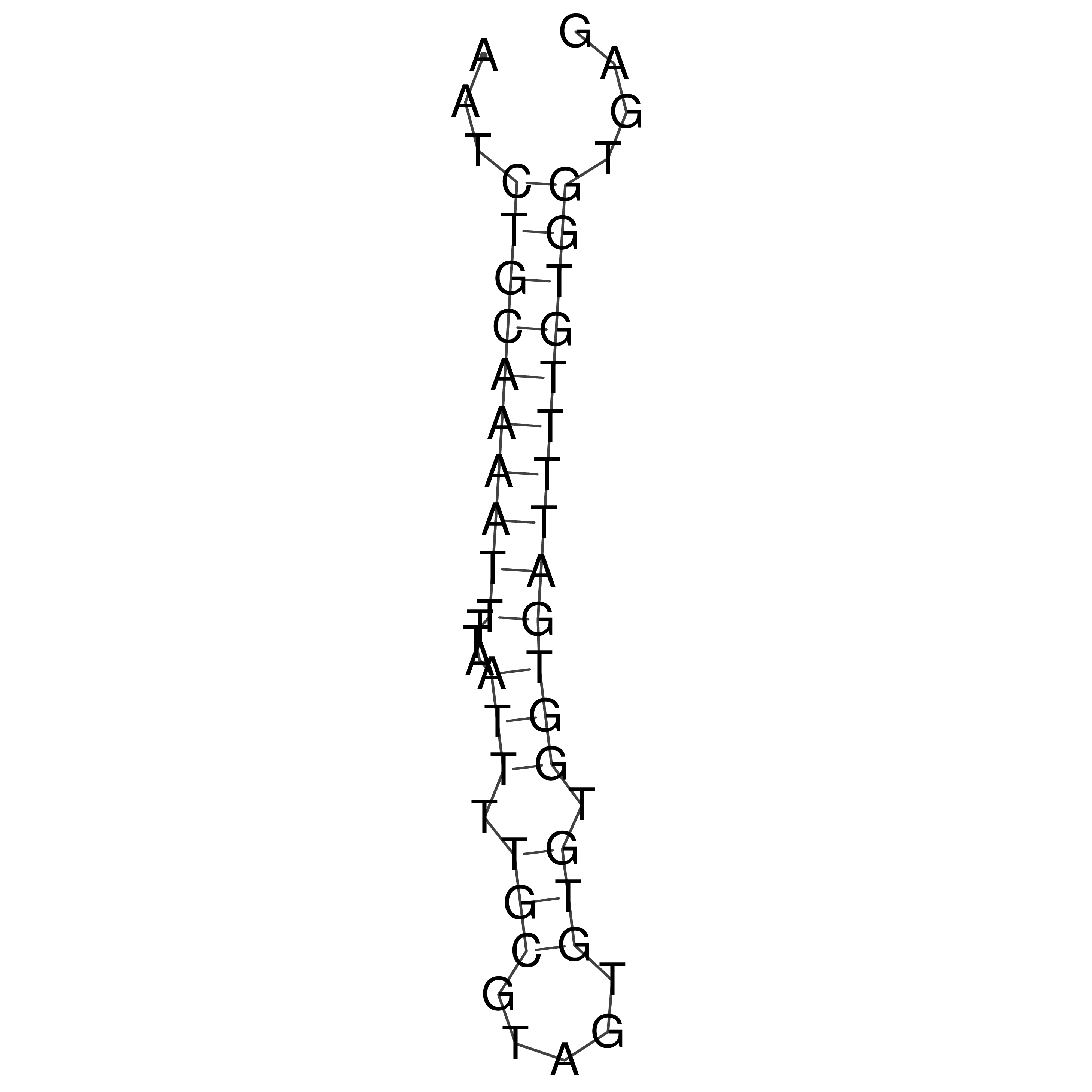
Relaxase
| ID | 1474 | GenBank | WP_182189506 |
| Name | traI_HV116_RS28655_pRHBSTW-00189_4 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190481.00 Da Isoelectric Point: 5.9706
>WP_182189506.1 conjugative transfer relaxase/helicase TraI [Klebsiella grimontii]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVES
LGYRTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 589 | GenBank | WP_004197733 |
| Name | WP_004197733_pRHBSTW-00189_4 |
UniProt ID | A0A0E3KB00 |
| Length | 129 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14835.90 Da Isoelectric Point: 4.6012
>WP_004197733.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Bacteria]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0E3KB00 |
T4CP
| ID | 1151 | GenBank | WP_064343438 |
| Name | traD_HV116_RS28650_pRHBSTW-00189_4 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85584.66 Da Isoelectric Point: 5.3307
>WP_064343438.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 73764..103907
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV116_RS28435 (HV116_28440) | 68866..69216 | + | 351 | WP_004153414 | hypothetical protein | - |
| HV116_RS28440 (HV116_28445) | 69846..70199 | + | 354 | WP_004152748 | hypothetical protein | - |
| HV116_RS28445 (HV116_28450) | 70256..70603 | + | 348 | WP_004152749 | hypothetical protein | - |
| HV116_RS28450 (HV116_28455) | 70698..70844 | + | 147 | WP_004152750 | hypothetical protein | - |
| HV116_RS28455 (HV116_28460) | 70895..71728 | + | 834 | WP_004152751 | N-6 DNA methylase | - |
| HV116_RS28460 (HV116_28465) | 72548..73369 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| HV116_RS28465 (HV116_28470) | 73402..73731 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| HV116_RS28470 (HV116_28475) | 73764..74249 | - | 486 | WP_022631514 | transglycosylase SLT domain-containing protein | virB1 |
| HV116_RS28475 (HV116_28480) | 74682..75071 | + | 390 | WP_004197733 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV116_RS28480 (HV116_28485) | 75310..75984 | + | 675 | WP_004197737 | hypothetical protein | - |
| HV116_RS28485 (HV116_28490) | 76134..76352 | + | 219 | WP_172619300 | TraY domain-containing protein | - |
| HV116_RS28490 (HV116_28495) | 76402..76770 | + | 369 | WP_042946346 | type IV conjugative transfer system pilin TraA | - |
| HV116_RS28495 (HV116_28500) | 76784..77089 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| HV116_RS28500 (HV116_28505) | 77109..77675 | + | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| HV116_RS28505 (HV116_28510) | 77662..78402 | + | 741 | WP_032738531 | type-F conjugative transfer system secretin TraK | traK |
| HV116_RS28510 (HV116_28515) | 78402..79826 | + | 1425 | WP_042946342 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV116_RS28515 (HV116_28520) | 79819..80415 | + | 597 | WP_023340943 | conjugal transfer pilus-stabilizing protein TraP | - |
| HV116_RS28520 (HV116_28525) | 80438..81007 | + | 570 | WP_023340942 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV116_RS28525 (HV116_28530) | 81139..81549 | + | 411 | WP_020805613 | hypothetical protein | - |
| HV116_RS28530 (HV116_28535) | 81554..81838 | + | 285 | WP_023316401 | hypothetical protein | - |
| HV116_RS28535 (HV116_28540) | 81862..82080 | + | 219 | WP_023316402 | hypothetical protein | - |
| HV116_RS28540 (HV116_28545) | 82081..82392 | + | 312 | WP_023316403 | hypothetical protein | - |
| HV116_RS28545 (HV116_28550) | 82459..82863 | + | 405 | WP_042939116 | hypothetical protein | - |
| HV116_RS28550 (HV116_28555) | 82860..83150 | + | 291 | WP_023340940 | hypothetical protein | - |
| HV116_RS28555 (HV116_28560) | 83158..83556 | + | 399 | WP_074186017 | hypothetical protein | - |
| HV116_RS28560 (HV116_28565) | 83628..86267 | + | 2640 | WP_042946320 | type IV secretion system protein TraC | virb4 |
| HV116_RS28565 (HV116_28570) | 86264..86656 | + | 393 | WP_042946321 | type-F conjugative transfer system protein TrbI | - |
| HV116_RS28570 (HV116_28575) | 86656..87282 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| HV116_RS28575 (HV116_28580) | 87326..88285 | + | 960 | WP_029497356 | conjugal transfer pilus assembly protein TraU | traU |
| HV116_RS28580 (HV116_28585) | 88300..88848 | + | 549 | WP_022631518 | hypothetical protein | - |
| HV116_RS28585 (HV116_28590) | 89569..90171 | + | 603 | WP_022631520 | hypothetical protein | - |
| HV116_RS28590 (HV116_28595) | 90315..90962 | + | 648 | WP_039698503 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV116_RS28595 (HV116_28600) | 91021..92976 | + | 1956 | WP_049192820 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV116_RS28600 (HV116_28605) | 93008..93262 | + | 255 | WP_004195500 | conjugal transfer protein TrbE | - |
| HV116_RS28605 (HV116_28610) | 93240..93488 | + | 249 | WP_004152675 | hypothetical protein | - |
| HV116_RS28610 (HV116_28615) | 93501..93827 | + | 327 | WP_004194451 | hypothetical protein | - |
| HV116_RS28615 (HV116_28620) | 93848..94600 | + | 753 | WP_042946327 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV116_RS28620 (HV116_28625) | 94611..94850 | + | 240 | WP_009309875 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV116_RS28625 (HV116_28630) | 94822..95394 | + | 573 | WP_009309876 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV116_RS28630 (HV116_28635) | 95387..95815 | + | 429 | WP_004152689 | hypothetical protein | - |
| HV116_RS28635 (HV116_28640) | 95802..97172 | + | 1371 | WP_009309877 | conjugal transfer pilus assembly protein TraH | traH |
| HV116_RS28640 (HV116_28645) | 97172..100045 | + | 2874 | WP_009309878 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV116_RS28645 (HV116_28650) | 100780..101472 | + | 693 | WP_009309879 | hypothetical protein | - |
| HV116_RS28650 (HV116_28655) | 101598..103907 | + | 2310 | WP_064343438 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2187 | GenBank | NZ_JABXQY010000004 |
| Plasmid name | pRHBSTW-00189_4 | Incompatibility group | IncFIB |
| Plasmid size | 115072 bp | Coordinate of oriT [Strand] | 74322..74370 [-] |
| Host baterium | Klebsiella grimontii strain RHBSTW-00189 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |