Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101725 |
| Name | oriT_pRHBSTW-00904_3 |
| Organism | Klebsiella pneumoniae strain RHBSTW-00904 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JABXPA010000003 (56463..56512 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pRHBSTW-00904_3
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file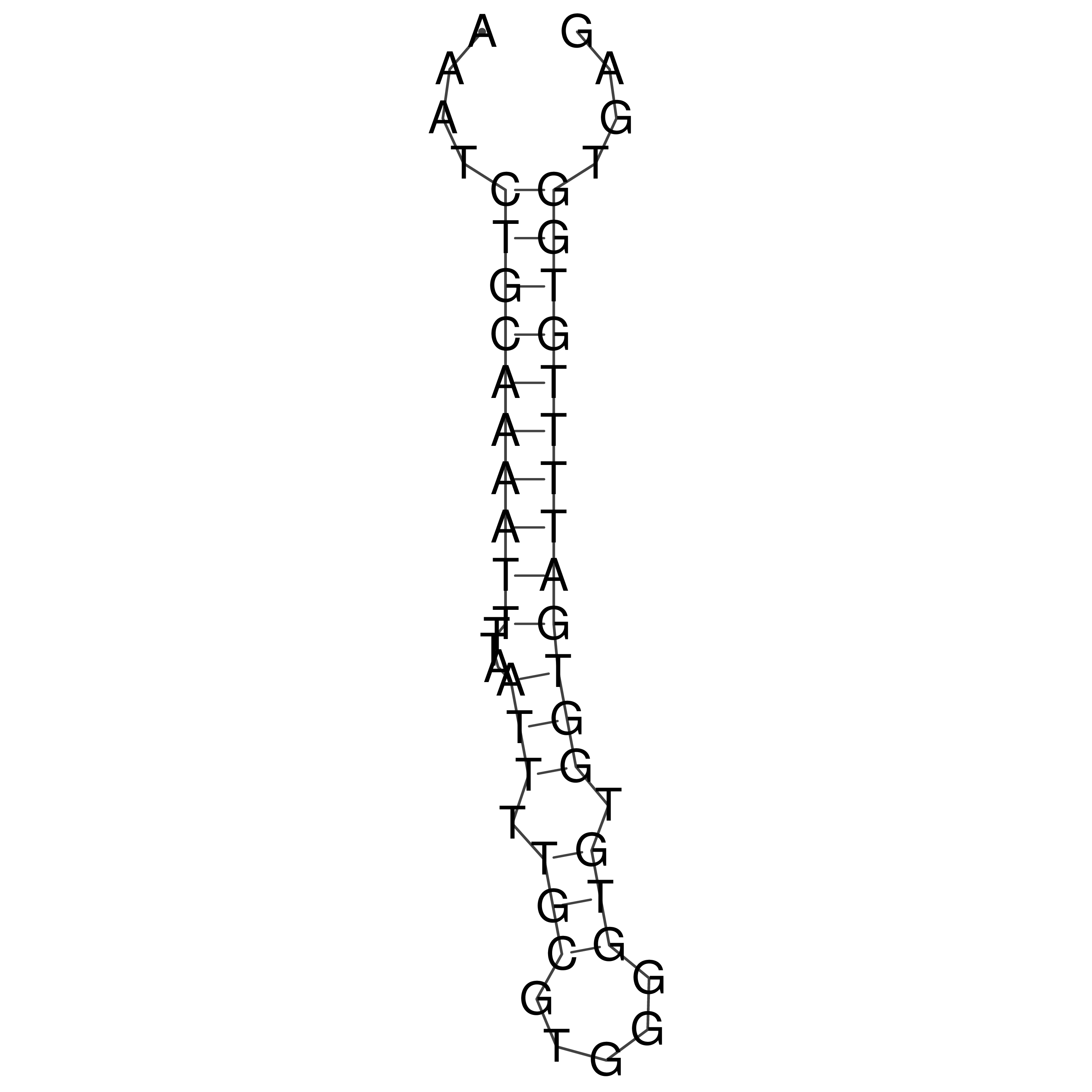
Relaxase
| ID | 1463 | GenBank | WP_065953842 |
| Name | traI_HV321_RS28070_pRHBSTW-00904_3 |
UniProt ID | A0A839CRE9 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190420.61 Da Isoelectric Point: 6.3863
>WP_065953842.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Klebsiella]
MMSIGSVKSADKAADYYTNKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADSRAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGLESVAQISGTREQSVLNGLIRDSLRNEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMESWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPAQEKAIQ
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHGARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQMITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARLLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
VDRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSADKAADYYTNKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRTQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADSRAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGLESVAQISGTREQSVLNGLIRDSLRNEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMESWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPAQEKAIQ
KALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHGARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQMITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILE
PRNDRAVKSADLLFGRARLLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
VDRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1142 | GenBank | WP_032436737 |
| Name | traD_HV321_RS28065_pRHBSTW-00904_3 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85553.49 Da Isoelectric Point: 5.0065
>WP_032436737.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREGRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGERPELASQPA
PAEVTVSPAPVKAPATTTMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREGRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGERPELASQPA
PAEVTVSPAPVKAPATTTMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 55905..85749
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV321_RS27845 (HV321_27845) | 52182..52538 | + | 357 | WP_004152717 | hypothetical protein | - |
| HV321_RS27850 (HV321_27850) | 52599..52811 | + | 213 | WP_004152718 | hypothetical protein | - |
| HV321_RS27855 (HV321_27855) | 52822..53046 | + | 225 | WP_004152719 | hypothetical protein | - |
| HV321_RS27860 (HV321_27860) | 53127..53447 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HV321_RS27865 (HV321_27865) | 53437..53715 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| HV321_RS27870 (HV321_27870) | 53716..54129 | + | 414 | WP_023280875 | transcriptional regulator | - |
| HV321_RS27875 (HV321_27875) | 54689..55516 | + | 828 | WP_023307552 | DUF932 domain-containing protein | - |
| HV321_RS27880 (HV321_27880) | 55543..55872 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| HV321_RS27885 (HV321_27885) | 55905..56390 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| HV321_RS27890 (HV321_27890) | 56825..57217 | + | 393 | WP_032436755 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV321_RS27895 (HV321_27895) | 57424..58119 | + | 696 | WP_023307551 | transcriptional regulator TraJ family protein | - |
| HV321_RS27900 (HV321_27900) | 58203..58574 | + | 372 | WP_004208838 | TraY domain-containing protein | - |
| HV321_RS27905 (HV321_27905) | 58628..58996 | + | 369 | WP_023307550 | type IV conjugative transfer system pilin TraA | - |
| HV321_RS27910 (HV321_27910) | 59010..59315 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| HV321_RS27915 (HV321_27915) | 59335..59901 | + | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| HV321_RS27920 (HV321_27920) | 59888..60628 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| HV321_RS27925 (HV321_27925) | 60628..62052 | + | 1425 | WP_023307549 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV321_RS27930 (HV321_27930) | 62045..62464 | + | 420 | Protein_76 | conjugal transfer pilus-stabilizing protein TraP | - |
| HV321_RS27935 (HV321_27935) | 62676..63245 | + | 570 | WP_023307547 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV321_RS27940 (HV321_27940) | 63377..63787 | + | 411 | WP_023307546 | hypothetical protein | - |
| HV321_RS27945 (HV321_27945) | 63792..64082 | + | 291 | WP_032423381 | hypothetical protein | - |
| HV321_RS27950 (HV321_27950) | 64106..64324 | + | 219 | WP_004171484 | hypothetical protein | - |
| HV321_RS27955 (HV321_27955) | 64325..64663 | + | 339 | WP_032436750 | hypothetical protein | - |
| HV321_RS27960 (HV321_27960) | 64708..65112 | + | 405 | WP_023307543 | hypothetical protein | - |
| HV321_RS27965 (HV321_27965) | 65109..65399 | + | 291 | WP_023307542 | hypothetical protein | - |
| HV321_RS27970 (HV321_27970) | 65407..65805 | + | 399 | WP_072158599 | hypothetical protein | - |
| HV321_RS27975 (HV321_27975) | 65877..68516 | + | 2640 | WP_016528865 | type IV secretion system protein TraC | virb4 |
| HV321_RS27980 (HV321_27980) | 68516..68905 | + | 390 | WP_032436749 | type-F conjugative transfer system protein TrbI | - |
| HV321_RS27985 (HV321_27985) | 68905..69531 | + | 627 | WP_023307538 | type-F conjugative transfer system protein TraW | traW |
| HV321_RS27990 (HV321_27990) | 69575..70534 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| HV321_RS27995 (HV321_27995) | 70547..71185 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV321_RS28000 (HV321_28000) | 71244..73199 | + | 1956 | WP_181467678 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV321_RS28005 (HV321_28005) | 73231..73467 | + | 237 | WP_032436746 | conjugal transfer protein TrbE | - |
| HV321_RS30475 | 73464..73649 | + | 186 | WP_032436745 | hypothetical protein | - |
| HV321_RS28010 (HV321_28010) | 73695..74021 | + | 327 | WP_032436744 | hypothetical protein | - |
| HV321_RS28015 (HV321_28015) | 74042..74794 | + | 753 | WP_015065637 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV321_RS28020 (HV321_28020) | 74805..75044 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV321_RS28025 (HV321_28025) | 75016..75573 | + | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV321_RS28030 (HV321_28030) | 75618..76060 | + | 443 | Protein_97 | F-type conjugal transfer protein TrbF | - |
| HV321_RS28035 (HV321_28035) | 76038..77417 | + | 1380 | WP_072158600 | conjugal transfer pilus assembly protein TraH | traH |
| HV321_RS28040 (HV321_28040) | 77417..79552 | + | 2136 | Protein_99 | conjugal transfer mating-pair stabilization protein TraG | - |
| HV321_RS28050 (HV321_28050) | 80785..81471 | + | 687 | Protein_101 | conjugal transfer protein TraG | - |
| HV321_RS28055 (HV321_28055) | 81495..82097 | + | 603 | WP_023280878 | hypothetical protein | - |
| HV321_RS28060 (HV321_28060) | 82349..83080 | + | 732 | WP_032415736 | conjugal transfer complement resistance protein TraT | - |
| HV321_RS30200 | 83271..83384 | + | 114 | Protein_104 | hypothetical protein | - |
| HV321_RS28065 (HV321_28065) | 83440..85749 | + | 2310 | WP_032436737 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 2169 | GenBank | NZ_JABXPA010000003 |
| Plasmid name | pRHBSTW-00904_3 | Incompatibility group | IncFIB |
| Plasmid size | 96966 bp | Coordinate of oriT [Strand] | 56463..56512 [-] |
| Host baterium | Klebsiella pneumoniae strain RHBSTW-00904 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merC, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |