Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101712 |
| Name | oriT_pRHBSTW-00265_4 |
| Organism | Escherichia marmotae strain RHBSTW-00265 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JABXQN010000004 (30290..30412 [+], 123 nt) |
| oriT length | 123 nt |
| IRs (inverted repeats) | 100..105, 118..123 (TTTAAT..ATTAAA) 90..98, 112..120 (AATAATGTA..TACATTATT) 89..94, 106..111 (AAATAA..TTATTT) 87..92, 104..109 (ATAAAT..ATTTAT) 38..45, 48..55 (GCAAAAAC..GTTTTTGC) 2..9, 14..21 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 123 nt
GTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTGGTTTATGTTATAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file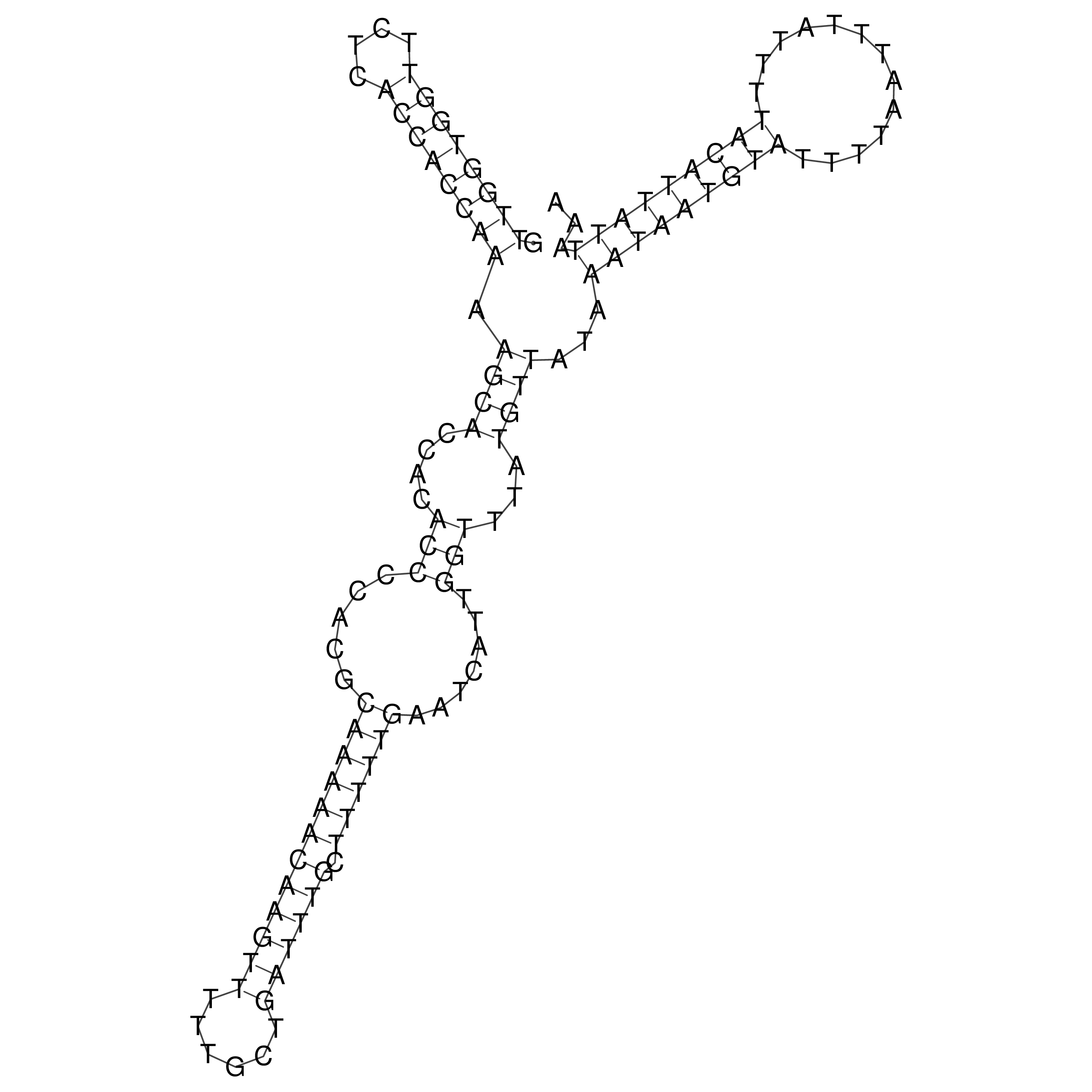
Relaxase
| ID | 1459 | GenBank | WP_182140475 |
| Name | traI_HV138_RS23340_pRHBSTW-00265_4 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191901.45 Da Isoelectric Point: 5.8177
MLSISVVKSAGSAGNYYTDKDNYYVLGSMEERWAGRGAEQLGLQGSVDKDVFTRLLEGKLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTLAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQILITDSGQRTGTGSALMAMKDAGVNIYRW
QGGEQRPATIISEPDRNVRYARLAEDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRQEKMPVADGERLRVTGKIPGLRVSGGDRLQVASTSEGSMTVVVPGRAEPATLPVGDSPFTALKL
ESGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLTRHPSFTVVSEQI
KTRAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSHVDLLTEAKSFATEGTSFADLGQEIDA
QIKRGDLLHVDVAKGYGTDLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQIM
VPVLSTANIRDGELRRLSTWETHRDALVLVDNVYHRIAGISKGDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGLQTRVIRPGQEQAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGGRKQMAGFESAYVALSRMKQHVQVYTDDRQGWTDAINNAVQKGTAHDVL
EPKADREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDISDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVREIAGQERERAAITEREAALPESVLRESQREREAIREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 582 | GenBank | WP_059222172 |
| Name | WP_059222172_pRHBSTW-00265_4 |
UniProt ID | _ |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14581.61 Da Isoelectric Point: 5.0359
MARVNLYISNEVHEKINMIVEKRRQEGARDKDISLSGTASMLLELGLRVYDAQVERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYTNMVDDIREKVSVEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 583 | GenBank | WP_071645896 |
| Name | WP_071645896_pRHBSTW-00265_4 |
UniProt ID | _ |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9005.06 Da Isoelectric Point: 9.7727
MRRRNARGGISRTVSVYLDEDTNNRLIQAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVTEESES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 1136 | GenBank | WP_182140459 |
| Name | traC_HV138_RS23240_pRHBSTW-00265_4 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99148.88 Da Isoelectric Point: 6.0798
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVKASWLAKKNQARIDDVVNFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1137 | GenBank | WP_182140473 |
| Name | traD_HV138_RS23335_pRHBSTW-00265_4 |
UniProt ID | _ |
| Length | 729 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 729 a.a. Molecular weight: 82916.64 Da Isoelectric Point: 5.1730
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKEVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVSTGKSEVIRRLANYAR
KRGDMVVIYDRSCEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNVANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTYLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDKKNGWLFISSNADTHASLKPVVSMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVLNTRAFFRSPSHQIA
EFAAGEIGEKEHLKASLQYSYGADPVRDGISTGKEMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPREINPEMENRLSAVLAAREAEGRQVASLFEPDVPEVVSGEDVVQAEQPQQPQQPQQ
PQQPQQPVSSVINDKKSDAGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPD
TQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 32665..52983
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV138_RS24200 | 27962..28168 | + | 207 | WP_249423634 | single-stranded DNA-binding protein | - |
| HV138_RS23165 (HV138_23165) | 28192..28488 | + | 297 | WP_182140510 | hypothetical protein | - |
| HV138_RS23170 (HV138_23170) | 28598..29419 | + | 822 | WP_182140512 | DUF932 domain-containing protein | - |
| HV138_RS23175 (HV138_23175) | 29717..30286 | - | 570 | Protein_48 | transglycosylase SLT domain-containing protein | - |
| HV138_RS23180 (HV138_23180) | 30642..31025 | + | 384 | WP_059222172 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV138_RS23185 (HV138_23185) | 31217..31903 | + | 687 | WP_182140514 | PAS domain-containing protein | - |
| HV138_RS23190 (HV138_23190) | 31997..32224 | + | 228 | WP_071645896 | conjugal transfer relaxosome protein TraY | - |
| HV138_RS23195 (HV138_23195) | 32258..32650 | + | 393 | WP_170898794 | type IV conjugative transfer system pilin TraA | - |
| HV138_RS23200 (HV138_23200) | 32665..32976 | + | 312 | WP_000012118 | type IV conjugative transfer system protein TraL | traL |
| HV138_RS23205 (HV138_23205) | 32998..33564 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| HV138_RS23210 (HV138_23210) | 33551..34279 | + | 729 | WP_016233172 | type-F conjugative transfer system secretin TraK | traK |
| HV138_RS23215 (HV138_23215) | 34279..35727 | + | 1449 | WP_182140516 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV138_RS23220 (HV138_23220) | 35717..36304 | + | 588 | WP_182140455 | conjugal transfer pilus-stabilizing protein TraP | - |
| HV138_RS23225 (HV138_23225) | 36291..36611 | + | 321 | WP_045896528 | conjugal transfer protein TrbD | - |
| HV138_RS23230 (HV138_23230) | 36608..37123 | + | 516 | WP_045896530 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV138_RS23235 (HV138_23235) | 37258..37479 | + | 222 | WP_182140457 | conjugal transfer protein TraR | - |
| HV138_RS23240 (HV138_23240) | 37639..40266 | + | 2628 | WP_182140459 | type IV secretion system protein TraC | virb4 |
| HV138_RS23245 (HV138_23245) | 40263..40649 | + | 387 | WP_000214093 | type-F conjugative transfer system protein TrbI | - |
| HV138_RS23250 (HV138_23250) | 40646..41278 | + | 633 | WP_045896538 | type-F conjugative transfer system protein TraW | traW |
| HV138_RS23255 (HV138_23255) | 41275..42267 | + | 993 | WP_045896540 | conjugal transfer pilus assembly protein TraU | traU |
| HV138_RS23260 (HV138_23260) | 42421..42789 | + | 369 | WP_045896542 | hypothetical protein | - |
| HV138_RS23265 (HV138_23265) | 42951..43286 | + | 336 | WP_182140461 | hypothetical protein | - |
| HV138_RS23270 (HV138_23270) | 43279..43731 | + | 453 | WP_170898601 | hypothetical protein | - |
| HV138_RS23275 (HV138_23275) | 43759..44397 | + | 639 | WP_170898603 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV138_RS23280 (HV138_23280) | 44394..46202 | + | 1809 | WP_182140463 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV138_RS23285 (HV138_23285) | 46229..46486 | + | 258 | WP_157940592 | conjugal transfer protein TrbE | - |
| HV138_RS23290 (HV138_23290) | 46479..47222 | + | 744 | WP_170898607 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV138_RS23295 (HV138_23295) | 47383..47619 | + | 237 | WP_212768171 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV138_RS23300 (HV138_23300) | 47606..48151 | + | 546 | WP_182140465 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV138_RS23305 (HV138_23305) | 48081..48422 | + | 342 | WP_182140467 | P-type conjugative transfer protein TrbJ | - |
| HV138_RS23310 (HV138_23310) | 48409..48801 | + | 393 | WP_249423636 | F-type conjugal transfer protein TrbF | - |
| HV138_RS23315 (HV138_23315) | 48788..50161 | + | 1374 | WP_105266958 | conjugal transfer pilus assembly protein TraH | traH |
| HV138_RS23320 (HV138_23320) | 50158..52983 | + | 2826 | WP_182140471 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV138_RS23325 (HV138_23325) | 52980..53489 | + | 510 | WP_000628099 | conjugal transfer entry exclusion protein TraS | - |
| HV138_RS23330 (HV138_23330) | 53503..54234 | + | 732 | WP_000850422 | conjugal transfer complement resistance protein TraT | - |
| HV138_RS23335 (HV138_23335) | 54487..56676 | + | 2190 | WP_182140473 | type IV conjugative transfer system coupling protein TraD | - |
Host bacterium
| ID | 2156 | GenBank | NZ_JABXQN010000004 |
| Plasmid name | pRHBSTW-00265_4 | Incompatibility group | - |
| Plasmid size | 70794 bp | Coordinate of oriT [Strand] | 30290..30412 [+] |
| Host baterium | Escherichia marmotae strain RHBSTW-00265 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF11 |