Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 101642 |
| Name | oriT_pMCR-E2899 |
| Organism | Escherichia coli strain MSHS 472 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_JABFOO010000006 (51677..51765 [-], 89 nt) |
| oriT length | 89 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 89 nt
>oriT_pMCR-E2899
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
GGGGTGTCGGGGCGAAGCCCTGACCAGATGGCAATTGTAATAGCGTCGCGTGTGACGGTATTACAATTGCACATCCTGTCCCGTTTTTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file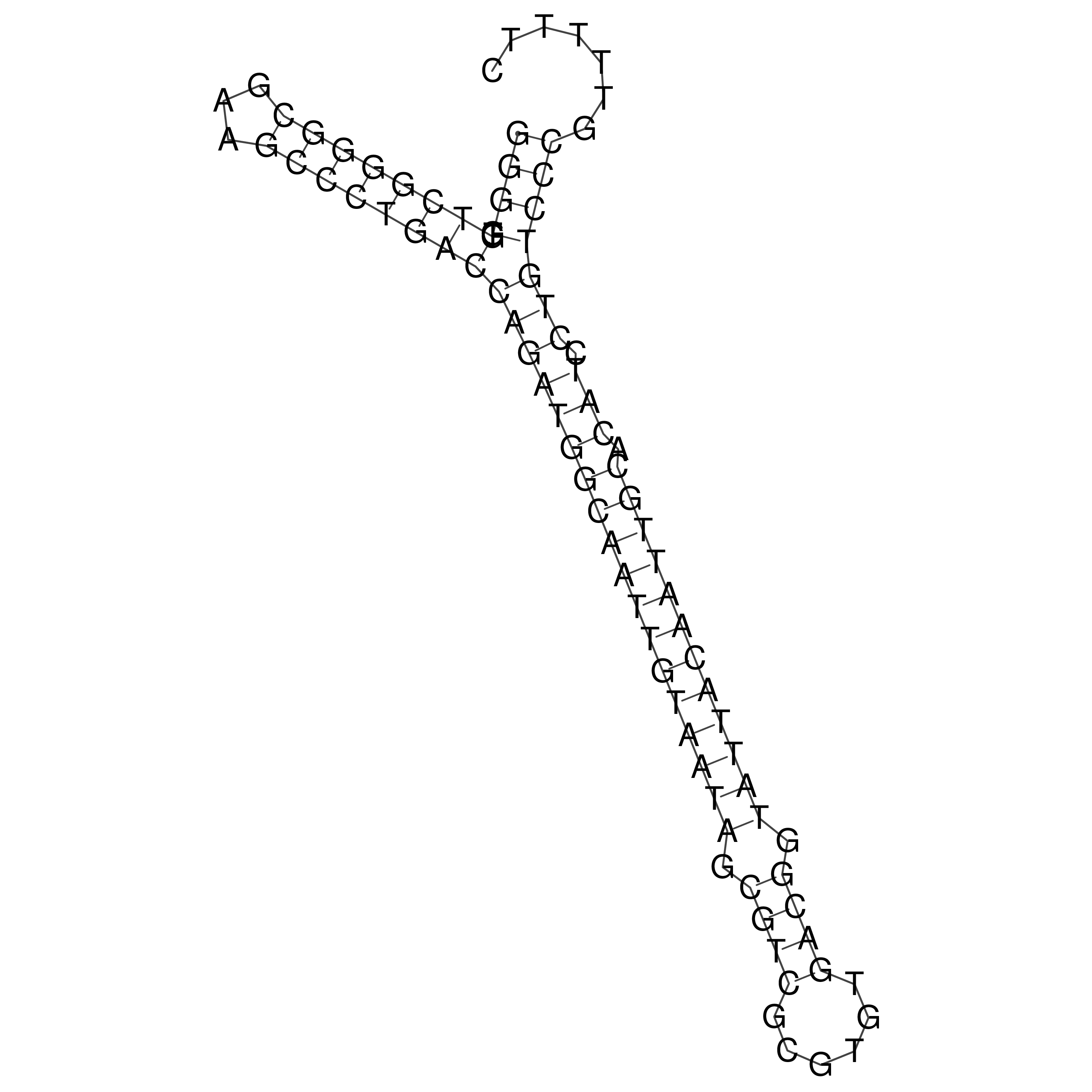
Relaxase
| ID | 1432 | GenBank | WP_052938718 |
| Name | nikB_HN009_RS22795_pMCR-E2899 |
UniProt ID | A0A706EBF4 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104042.50 Da Isoelectric Point: 7.3623
>WP_052938718.1 MULTISPECIES: IncI1-type relaxase NikB [Enterobacteriaceae]
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLAERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
MNAVIPKKRRDGKSSFEDLVSYVSVRDDMTDEELDLSSSSQAEQPHRSRFSRLVDYATRLRNESFVALVD
VMKDGCEWVNFYGVTCFHNCTSLETAAADMEYIARQAHYAKDDTDPVFHYILSWQSHESPRPEQIYDSVR
HTLKSLGLADHQYVSAVHTDTDNLHVHVAVNRVHPETGYLNRLSWSQEKLSRACRELELKHGFAPDNGCW
VHAPGNRIVRKTAVERDRQNAWTRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSHMDGK
FLVMDGWDRNREGVQLDSFGPSWCAEKLMKKMGDYTPVPKDIFSQVEAPGRYNPDFIAADVRPEKIAETE
SLQQYACRHLAERLPEMAREGRLENCQAIHRTLAEAGLWMRVQHGHLVICDGYDHNQTPVRADSVWSLLT
LDNVNQLDGGWLPVPTDIFRQVTPTERFRGRRMESCPATDKEWHRMRTGTGPQGAIKRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLGRAEP
QAGPFVSAPADLFDRVQPESRYNPELAVSDRYGVSSKRDPMLRRQRREARAEARADLRARYLAWREQWRK
PDLRYGERCREIHQACRLRKSHIRAQYDDPALRKLHYHIAEVQRMQALIRLKEDIRDERQKLIADGKWYP
PSYRQWVEIQAAQGDRAAVSQLRGWDYRDRRKDRSRTTTTDRCVVLCEPGGTPVYGNTGDLEARLQKNGS
VRFRDRRTGEFVCTDYGDRVVFRNHHDRNALADKLDLIAPVLFGRDPRMGFEPEGNDKQFNQVFAEMVAW
HNVTGRTGHEDYRITRPDVDHHREGSERYYRDYIAANSNDDASLPPPEQDKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A706EBF4 |
Auxiliary protein
| ID | 563 | GenBank | WP_052938717 |
| Name | WP_052938717_pMCR-E2899 |
UniProt ID | A0A706B4V0 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12643.59 Da Isoelectric Point: 10.7463
>WP_052938717.1 MULTISPECIES: IncI1-type relaxosome accessory protein NikA [Enterobacteriaceae]
MSDSTVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
MSDSTVRKKSEVRQKTVVRTLRFSPVEDETIRKKAEDSGLTVSAYIRNAALNKRINSRTDDAFLKELMRL
GRMQKHLFVQGKRTGDKEYAEVLVAITELTNTLRKQLMEG
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A706B4V0 |
T4CP
| ID | 1098 | GenBank | WP_021497839 |
| Name | trbC_HN009_RS22800_pMCR-E2899 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86967.19 Da Isoelectric Point: 7.0662
>WP_021497839.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLKCADPSQDRMIKRSLFSFWPTLFQYEVILESPANGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKTNRPDWDGMY
MSEHRVNPELLHRTAWGNPVWNALQSLNIYGFCLVASLVASFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLKCADPSQDRMIKRSLFSFWPTLFQYEVILESPANGIFYVGYQRVRDIGRELWLSMDDLTRHIM
FFATTGGGKTETIFAWAINPLCWARGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDLSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTEAFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFITQKSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNNLQRQDGIFGESWIDSPQISILMESKINVQELIELH
PGEFFSIFRGETVPSASFFIPDDEKSCSSDPVVINRYISVDAPRLDRLRRLVPRTTQRRIPSPENVSAII
GVLTAKPSRKRRKIRTEPHTIVDTFQQRIAGRQAAMAMLEEYDTDINARESALWETAVNTLKTTTREERR
IRYITLNRPELPETKEENQISVRAERAGINLLTLPQDNNHPTGRPVNGFHHKKTNRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 57143..97636
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HN009_RS22800 (HN009_22805) | 54859..57150 | - | 2292 | WP_021497839 | F-type conjugative transfer protein TrbC | - |
| HN009_RS22805 (HN009_22810) | 57143..58213 | - | 1071 | WP_000151583 | IncI1-type conjugal transfer protein TrbB | trbB |
| HN009_RS22810 (HN009_22815) | 58232..59440 | - | 1209 | WP_000121273 | IncI1-type conjugal transfer protein TrbA | trbA |
| HN009_RS22815 (HN009_22820) | 59732..59884 | + | 153 | WP_001387489 | Hok/Gef family protein | - |
| HN009_RS22820 (HN009_22825) | 59956..60207 | - | 252 | WP_001291964 | hypothetical protein | - |
| HN009_RS22825 (HN009_22830) | 60527..62098 | - | 1572 | WP_171993024 | IS66-like element ISCro1 family transposase | - |
| HN009_RS22830 (HN009_22835) | 62118..62465 | - | 348 | WP_000624622 | IS66 family insertion sequence element accessory protein TnpB | - |
| HN009_RS22835 (HN009_22840) | 62465..63142 | - | 678 | WP_001682408 | IS66-like element accessory protein TnpA | - |
| HN009_RS24185 | 63413..63508 | + | 96 | WP_001303310 | DinQ-like type I toxin DqlB | - |
| HN009_RS22840 (HN009_22845) | 63573..63749 | - | 177 | WP_001054898 | hypothetical protein | - |
| HN009_RS22845 (HN009_22850) | 64149..64358 | + | 210 | WP_000062603 | HEAT repeat domain-containing protein | - |
| HN009_RS22850 (HN009_22855) | 64430..65080 | - | 651 | WP_001178506 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HN009_RS22855 (HN009_22860) | 65154..67322 | - | 2169 | WP_052938719 | DotA/TraY family protein | traY |
| HN009_RS22860 (HN009_22865) | 67419..68003 | - | 585 | WP_001476649 | IncI1-type conjugal transfer protein TraX | - |
| HN009_RS22865 (HN009_22870) | 68032..69234 | - | 1203 | WP_014065727 | IncI1-type conjugal transfer protein TraW | traW |
| HN009_RS22870 (HN009_22875) | 69201..69815 | - | 615 | WP_000337398 | IncI1-type conjugal transfer protein TraV | traV |
| HN009_RS22875 (HN009_22880) | 69815..72859 | - | 3045 | WP_001024782 | IncI1-type conjugal transfer protein TraU | traU |
| HN009_RS22880 (HN009_22885) | 72949..73749 | - | 801 | WP_021497829 | IncI1-type conjugal transfer protein TraT | traT |
| HN009_RS22885 (HN009_22890) | 73733..73921 | - | 189 | WP_001277255 | putative conjugal transfer protein TraS | - |
| HN009_RS22890 (HN009_22895) | 73985..74389 | - | 405 | WP_021497835 | IncI1-type conjugal transfer protein TraR | traR |
| HN009_RS22895 (HN009_22900) | 74440..74967 | - | 528 | WP_001055569 | conjugal transfer protein TraQ | traQ |
| HN009_RS22900 (HN009_22905) | 74967..75671 | - | 705 | WP_001493811 | IncI1-type conjugal transfer protein TraP | traP |
| HN009_RS22905 (HN009_22910) | 75671..76960 | - | 1290 | WP_001271991 | conjugal transfer protein TraO | traO |
| HN009_RS22910 (HN009_22915) | 76963..77946 | - | 984 | WP_171993026 | IncI1-type conjugal transfer protein TraN | traN |
| HN009_RS22915 (HN009_22920) | 77957..78649 | - | 693 | WP_000138548 | DotI/IcmL family type IV secretion protein | traM |
| HN009_RS22920 (HN009_22925) | 78646..78993 | - | 348 | WP_001055900 | conjugal transfer protein | traL |
| HN009_RS22925 (HN009_22930) | 79011..82778 | - | 3768 | WP_171993027 | LPD7 domain-containing protein | - |
| HN009_RS24230 | 82868..82951 | - | 84 | Protein_95 | phospholipase D family protein | - |
| HN009_RS22930 (HN009_22935) | 83039..83419 | + | 381 | WP_001171523 | transposase | - |
| HN009_RS22935 (HN009_22940) | 83416..83763 | + | 348 | WP_000612556 | IS66 family insertion sequence element accessory protein TnpB | - |
| HN009_RS22940 (HN009_22945) | 83859..85451 | - | 1593 | WP_001593684 | IS66 family transposase | - |
| HN009_RS22945 (HN009_22950) | 85482..85832 | - | 351 | WP_000624717 | IS66 family insertion sequence element accessory protein TnpB | - |
| HN009_RS22950 (HN009_22955) | 85829..86254 | - | 426 | WP_000422741 | transposase | - |
| HN009_RS22955 (HN009_22960) | 86269..86820 | - | 552 | WP_252409331 | phospholipase D family protein | - |
| HN009_RS22960 (HN009_22965) | 86835..87125 | - | 291 | WP_001299214 | hypothetical protein | traK |
| HN009_RS22965 (HN009_22970) | 87122..88270 | - | 1149 | WP_001024972 | plasmid transfer ATPase TraJ | virB11 |
| HN009_RS22970 (HN009_22975) | 88267..89085 | - | 819 | WP_032220737 | IncI1-type conjugal transfer lipoprotein TraI | traI |
| HN009_RS22975 (HN009_22980) | 89082..89540 | - | 459 | WP_001079808 | IncI1-type conjugal transfer lipoprotein TraH | - |
| HN009_RS24060 | 89935..90096 | - | 162 | Protein_106 | histidine phosphatase family protein | - |
| HN009_RS22985 (HN009_22990) | 91606..91854 | + | 249 | WP_001349157 | hypothetical protein | - |
| HN009_RS22990 (HN009_22995) | 91851..93143 | - | 1293 | WP_001619585 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HN009_RS22995 (HN009_23000) | 93143..93799 | - | 657 | WP_001193549 | A24 family peptidase | - |
| HN009_RS23000 (HN009_23005) | 93784..94344 | - | 561 | WP_000014005 | lytic transglycosylase domain-containing protein | virB1 |
| HN009_RS23005 (HN009_23010) | 94354..94968 | - | 615 | WP_000908226 | type 4 pilus major pilin | - |
| HN009_RS23010 (HN009_23015) | 94985..96070 | - | 1086 | WP_001208802 | type II secretion system F family protein | - |
| HN009_RS23015 (HN009_23020) | 96083..97636 | - | 1554 | WP_000362206 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HN009_RS23020 (HN009_23025) | 97647..98099 | - | 453 | WP_001247337 | type IV pilus biogenesis protein PilP | - |
| HN009_RS23025 (HN009_23030) | 98086..99381 | - | 1296 | WP_000752780 | type 4b pilus protein PilO2 | - |
| HN009_RS23030 (HN009_23035) | 99374..101056 | - | 1683 | WP_000748143 | PilN family type IVB pilus formation outer membrane protein | - |
| HN009_RS23035 (HN009_23040) | 101070..101507 | - | 438 | WP_000539811 | type IV pilus biogenesis protein PilM | - |
| HN009_RS23040 (HN009_23045) | 101507..102574 | - | 1068 | WP_054170954 | type IV pilus biogenesis lipoprotein PilL | - |
Host bacterium
| ID | 2086 | GenBank | NZ_JABFOO010000006 |
| Plasmid name | pMCR-E2899 | Incompatibility group | IncI1 |
| Plasmid size | 107399 bp | Coordinate of oriT [Strand] | 51677..51765 [-] |
| Host baterium | Escherichia coli strain MSHS 472 |
Cargo genes
| Drug resistance gene | blaTEM-1B, mcr-1.1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |